Thayer Lab
Mathew J. Thayer, Ph.D.

Associate Professor
Department of Biochemistry and Molecular Biology
Oregon Health & Science University
3181 SW Sam Jackson Park Road
Portland OR 97239
(503)494-2447
thayerm@ohsu.edu
RESEARCH INTERESTS
Cancer cells differ from their normal cellular counterparts in many important characteristics, including loss of differentiation, increased genomic instability, and decreased drug sensitivity. Not surprisingly, genetic alterations occur in most, if not all cancer cells, and are thought to lie at the heart of these phenotypic alterations. Furthermore, genetic instability is thought to be required to generate the multiple genetic changes that occur in cancer cells. My laboratory uses somatic cell and molecular genetics to identify and characterize genetic alterations found in tumor cells that induce abnormal cellular phenotypes. By utilizing this approach, my lab has identified a previously unknown chromosomal abnormality that is associated with certain chromosomal rearrangements. This chromosomal phenotype is characterized by a delay in mitotic chromosome condensation, a delay in the chromosome replication timing, and significant chromosomal instability. Chromosomes with this phenotype are common in tumor derived cell lines and in primary tumors. Furthermore, we have found that exposing cells to ionizing radiation generates chromosomes with this phenotype. Our findings support a model in which the chromosomal instability found in tumor cells, and in cells exposed to ionizing radiation, stems from a defect in the replication timing of certain chromosomal rearrangements. Recently, we developed a chromosome engineering strategy that allows us to generate chromosomes with this delayed replication and condensation phenotype in an efficient and reproducible manner. Our findings indicate that ~5% of all random chromosome translocations display this abnormal phenotype. In addition, on certain balanced translocations only one of the derivative chromosomes displays the phenotype, indicating that a cis-acting mechanism is responsible for this abnormal chromosomal phenotype. The primary goal of this proposal is to characterize this cis-acting mechanism that functions to delay the replication timing of entire chromosomes. This proposal utilizes ‘chromosome engineering’ strategies, combined with somatic cell and molecular genetic approaches, to generate and characterize chromosomes with this delayed replication and condensation phenotype. The long-term goal of these studies is to define the molecular mechanisms responsible for chromosomal instability, one of the most common types of genetic instabilities found in cancer cells.
Chromosomes with DRT/DMC
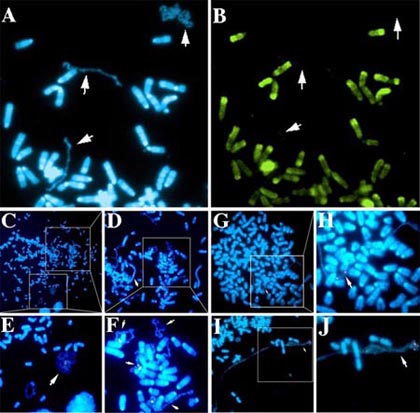
My lab has characterized an abnormal chromosomal phenotype that is associated with certain tumor-derived chromosome alterations. These chromosomes display a significant delay in mitotic chromosome condensation (DMC), associated with a delay in the mitosis-specific phosphorylation of histone H3, and a 2-3 hour delay in the replication timing (DRT) of the entire chromosome. In addition, chromosomes with DRT/DMC participate in frequent secondary translocations and rearrangements, indicating that cells containing these chromosomes display CIN.
Individual cells can contain multiple chromosomes with the DMC phenotype, and the DMC chromosomes within a given cell can display a wide range in the extent of mitotic chromosome condensation (Fig. 1C-F). Furthermore, chromosomes with the more extreme under-condensed appearance lack the mitosis-specific phosphorylation of histone H3 on serine 10 (Fig. 1A and B) usually initiated in late G2 and completed just prior to nuclear envelope breakdown and the formation of prophase chromosomes. These observations suggest that the more extreme under-condensed chromosomes retain an “interphase state” of condensation in cells that contain fully phosphorylated and condensed metaphase chromosomes.
Engineering DRT/DMC
We have developed a chromosome engineering system that allows us to: 1) target the genome in a random fashion, 2) create reciprocal chromosome translocations at defined locations, 3) generate the same translocations in multiple independent events using two distinct mechanisms, and 4) characterize the chromosomes both before and after the translocation events. The chromosome engineering strategy that we are employing is based on the Cre/loxP site-specific recombinase system to generate inter-chromosomal translocations. Because the Cre/loxP system is relatively inefficient at generating inter-chromosomal translocations (<1 x 10-3), we are using reconstitution of a selectable marker to isolate the cells that undergo Cre-mediated recombination. A schematic representation of this strategy is shown in Fig. 2.