Newman Lab: Difference between revisions
Yuanrandong (talk | contribs) No edit summary |
Yuanrandong (talk | contribs) No edit summary |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<p class=MsoTitle style='border:none'><span style='font-size:12.0pt;font-family: | |||
"Times New Roman"'><big>'''Curriculum Vitae'''</big></span></p> | |||
<p class=MsoTitle style='border:none'><span style='font-size: | |||
"Times New Roman"'>Curriculum Vitae</span></p> | |||
<p class=MsoTitle style='border:none'><span style='font-size:10.0pt;font-family: | <p class=MsoTitle style='border:none'><span style='font-size:10.0pt;font-family: | ||
| Line 165: | Line 35: | ||
APPOINTMENTS</span></u></b><u><span style='font-size:10.0pt'>:</span></u></p> | APPOINTMENTS</span></u></b><u><span style='font-size:10.0pt'>:</span></u></p> | ||
<p class=MsoNormal><span style='font-size:10.0pt'>2006.6 - | <p class=MsoNormal><span style='font-size:10.0pt'>2006.6 - Now Assistant | ||
Professor, Nanyang Technological University, Singapore</span></p> | Professor, Nanyang Technological University, Singapore</span></p> | ||
| Line 343: | Line 213: | ||
to study their binding kinetics.</span></p> | to study their binding kinetics.</span></p> | ||
<p> | <p>[[image:protein.gif]]</p> | ||
[[image: | |||
<p class=MsoNormal><span style='font-size:9.0pt;font-family:Verdana;color:black'>In | <p class=MsoNormal><span style='font-size:9.0pt;font-family:Verdana;color:black'>In | ||
| Line 369: | Line 238: | ||
Intensity Descent Method (AID-HRMS) for Efficient Interpretation of Complex | Intensity Descent Method (AID-HRMS) for Efficient Interpretation of Complex | ||
High Resolution Mass Spectra of Proteins, </i>Analytical Chemistry<i>, </i>2006. | High Resolution Mass Spectra of Proteins, </i>Analytical Chemistry<i>, </i>2006. | ||
(In Press). | (In Press).</span></p> | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
| Line 380: | Line 249: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'>3.<span style='font:7.0pt "Times New Roman"'> | style='font-size:10.0pt'>3.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Guo X, Chan-Park M, Yoon SF, Chun | </span></span><span style='font-size:10.0pt'>Guo X, Chan-Park M, Yoon SF, Chun | ||
J, Hua L, and Sze NSK, <i>UV Embossed Polymeric Chip for Protein Separation and | J, Hua L, and Sze NSK, <i>UV Embossed Polymeric Chip for Protein Separation and | ||
| Line 394: | Line 255: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>4.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Hua L, Low TY, and Sze NSK, <i>Microwave-assisted | </span></span><span style='font-size:10.0pt'>Hua L, Low TY, and Sze NSK, <i>Microwave-assisted | ||
Specific Chemical Digestion for Rapid Protein Identification, </i>Proteomics, <b>6</b>, | Specific Chemical Digestion for Rapid Protein Identification, </i>Proteomics, <b>6</b>, | ||
| Line 400: | Line 261: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>5.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Aye TT, Low TY, and Sze NSK, <i>Nanosecond | </span></span><span style='font-size:10.0pt'>Aye TT, Low TY, and Sze NSK, <i>Nanosecond | ||
Laser Induced Photochemical Oxidation method for Protein Surface Mapping with | Laser Induced Photochemical Oxidation method for Protein Surface Mapping with | ||
| Line 406: | Line 267: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>6.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Mok MLS, Hua Lin, Phua JBC, Wee | </span></span><span style='font-size:10.0pt'>Mok MLS, Hua Lin, Phua JBC, Wee | ||
MKT, and Sze NSK, <i>Capillary Isoelectric Focusing in Pseudo-closed Channel | MKT, and Sze NSK, <i>Capillary Isoelectric Focusing in Pseudo-closed Channel | ||
| Line 413: | Line 274: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>7.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Sze NSK, Ge Y, Oh H, and McLafferty | </span></span><span style='font-size:10.0pt'>Sze NSK, Ge Y, Oh H, and McLafferty | ||
FW, <i>Plasma Electron Capture Dissociation for Large Protein</i>, Analytical | FW, <i>Plasma Electron Capture Dissociation for Large Protein</i>, Analytical | ||
| Line 419: | Line 280: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>8.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Ge Y, ElNaggar M, Sze NSK, Hanbin | </span></span><span style='font-size:10.0pt'>Ge Y, ElNaggar M, Sze NSK, Hanbin | ||
Oh, Begley TP, and McLafferty FW, <i>Top Down Proteomic Analysis of Secreted | Oh, Begley TP, and McLafferty FW, <i>Top Down Proteomic Analysis of Secreted | ||
| Line 427: | Line 288: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>9.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Ge Y, Lwahor BG, ElNaggar M, Sze | </span></span><span style='font-size:10.0pt'>Ge Y, Lwahor BG, ElNaggar M, Sze | ||
NSK, Begley TP, and McLafferty FW, <i>Protein Oxidation in Viral | NSK, Begley TP, and McLafferty FW, <i>Protein Oxidation in Viral | ||
| Line 434: | Line 295: | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>10.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Oh H, Breuker K, Sze SK, Ge Y, | </span></span><span style='font-size:10.0pt'>Oh H, Breuker K, Sze SK, Ge Y, | ||
Carpenter BK, and McLafferty FW, <i>Secondary and tertiary structures of | Carpenter BK, and McLafferty FW, <i>Secondary and tertiary structures of | ||
gaseous protein ions characterized by electron capture dissociation mass | gaseous protein ions characterized by electron capture dissociation mass | ||
spectrometry and photofragment spectroscopy.</i | spectrometry and photofragment spectroscopy.</i> Proceedings Of The National Academy Of Sciences Of The United States Of America, <b>99, | ||
Of The | |||
</b>15863-8, 2002.</span></p> | </b>15863-8, 2002.</span></p> | ||
<p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.25in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'> | style='font-size:10.0pt'>11.<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>Sze NSK, Ge Y, Oh H, and McLafferty | </span></span><span style='font-size:10.0pt'>Sze NSK, Ge Y, Oh H, and McLafferty | ||
FW, <i>Top Down Mass Spectrometry of a 29 kDa Protein for Characterization of | FW, <i>Top Down Mass Spectrometry of a 29 kDa Protein for Characterization of | ||
| Line 492: | Line 352: | ||
<p class=MsoNormal><span style='font-size:10.0pt'>NARDEV S/O RAMANATHAN</span></p> | <p class=MsoNormal><span style='font-size:10.0pt'>NARDEV S/O RAMANATHAN</span></p> | ||
<p class=MsoNormal><span style='font-size:10.0pt'>Email | <p class=MsoNormal><span style='font-size:10.0pt'>Email: nardev@pmail.ntu.edu.sg</span></p> | ||
<p class=MsoNormal><span style='font-size:10.0pt'>Tel: (65) 6316-2852</span></p> | <p class=MsoNormal><span style='font-size:10.0pt'>Tel: (65) 6316-2852</span></p> | ||
| Line 500: | Line 359: | ||
<p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | <p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | ||
<p class=MsoNormal><span style='font-size:10.0pt'>Zhao Chen</span></p> | |||
<p class=MsoNormal><span style='font-size:10.0pt'>Email: 149914862@ntu.edu.sg</span></p> | |||
<p class=MsoNormal><span style='font-size:10.0pt'>Tel: (65) 6316-2852</span></p> | |||
<p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | |||
<p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | |||
<p class=MsoNormal><b><u><span style='font-size:10.0pt'>Vacancies:</span></u></b></p> | <p class=MsoNormal><b><u><span style='font-size:10.0pt'>Vacancies:</span></u></b></p> | ||
| Line 505: | Line 375: | ||
<p class=MsoNormal><span style='font-size:10.0pt'>We are looking for | <p class=MsoNormal><span style='font-size:10.0pt'>We are looking for | ||
Postdoctoral Fellows, Research Assistants and Graduate Students to join our | Postdoctoral Fellows, Research Assistants and Graduate Students to join our | ||
multi-disciplinary team in the following | multi-disciplinary team in the following areas:</span></p> | ||
<p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | <p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | ||
| Line 512: | Line 382: | ||
style='font-size:10.0pt'>1)<span style='font:7.0pt "Times New Roman"'> | style='font-size:10.0pt'>1)<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>proteomics technology development: | </span></span><span style='font-size:10.0pt'>proteomics technology development: | ||
background in physics | background in physics, chemistry, biology or engineering</span></p> | ||
<p class=MsoNormal style='margin-left:.5in;text-indent:-.25in'><span | <p class=MsoNormal style='margin-left:.5in;text-indent:-.25in'><span | ||
style='font-size:10.0pt'>2)<span style='font:7.0pt "Times New Roman"'> | style='font-size:10.0pt'>2)<span style='font:7.0pt "Times New Roman"'> | ||
</span></span><span style='font-size:10.0pt'>proteomics bioinformatics/mass | </span></span><span style='font-size:10.0pt'>proteomics bioinformatics/mass | ||
spectrometry data analysis: Mathematics, Computer Sciences | spectrometry data analysis: Mathematics, Computer Sciences </span></p> | ||
<p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | <p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | ||
| Line 530: | Line 400: | ||
<p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | <p class=MsoNormal><span style='font-size:10.0pt'> </span></p> | ||
Latest revision as of 03:15, 8 September 2006
Curriculum Vitae
Name: Newman Siu Kwan SZE
Title: Assistant Professor, School of Biological Sciences, Nanyang Technological University
EMail: sksze@ntu.edu.sg
EDUCATION:
1990-1995 Ph.D., University of Hong Kong, Hong Kong
1987-1990 B.Sc., University of Hong Kong, Hong Kong
PROFESSIONAL APPOINTMENTS:
2006.6 - Now Assistant Professor, Nanyang Technological University, Singapore
2002.7 – 2006.5 Group Leader, Genome Institute of Singapore, Singapore
2003.7 – 2006.5 Adjunct Assistant Professor, National University of Singapore, Singapore
2005.5 – 2006.5 Adjunct Assistant Professor, Nanyang Technology University, Singapore
2001.5 – 2002.6 Visiting Scientist, Cornell University, USA
1998.6 – 2001.5 Postdoctoral Fellow, University of Waterloo, Canada
1995.5 – 1998.5 Postdoctoral Fellow, University of Toronto, Canada
HONORS AND AWARDS:
1. Human Proteome Organization Annual Congress - First Prize Poster Award, 2004
Probing protein solvent accessible surface by laser induced oxidation and mass spectrometry
2. The Croucher Foundation Fellowships, 1995-97
3. The Croucher Foundation Studentships, 1990-93
4. Sir Edward Youde Memorial Scholarships, 1989-90
5. Du pont Science Scholarships, 1989-90
6. Dow Chemical Scholarships, 1989-90
7. G.T. Byrne Memorial Prize in Chemistry, 1989-90
8. Douglas Payne Prizes in Chemistry, 1989-90
INVITED SEMINARS:
1. Workshop on Proteomics Data Analysis and Data Mining 2005
Algorithm for Quantitative Proteome Analysis
2. 5th HUGO Pacific Conference Nov 2004
Fourier Transform Mass Spectrometry for Top Down Proteomics
3. The Mass Spectrometry Society of Japan - The 52nd Annual Conference 2004
Application of Top Down Proteomics to Epigenetics and Gene Regulation
4. The Mass Spectrometry Society of Japan, Kanto-area Colloquium
Probing Protein Tertiary Structure and Interaction by Mass Spectrometry
5. Pharmacoproteomics 2003, Tokyo, Japan
Top Down Approach for Proteomics Analysis by Fourier Transform Mass Spectrometry and Electron Capture Dissociation
6. A Workshop on Introduction to BioMEMS and BioMedical Applications 2003, Institute of Microelectronics, Singapore
Coupling Capillary Isoelectric Focusing in Open Microchannel to Mass Spectrometry for Protein Characterization
7. Proteomics in Asia and Oceania 2002, Osaka, Japan
Fourier Transform Mass Spectrometry and Electron Capture Dissociation for Protein Characterization
SCIENTIFIC SERVICES:
1. Editorial Board, Journal of Mass Spectrometry Society of Japan
2. Proposal Reviewer, Hong Kong Research Grant Council
3. Proposal Reviewer, BMRC, Singapore
RESEARCH INTERESTS:
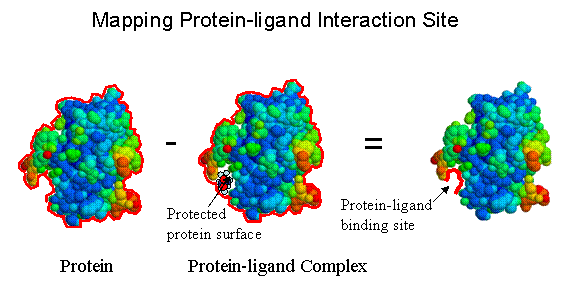
Our research is devoted to develop and apply advanced proteomic technologies to decipher human diseases through a rational search for novel disease targets and medicines. Tandem affinity purification tags have been constructed and utilized to study proteins that are important to transcriptional and translational regulatory networks, DNA repair, and apoptotic pathway. To this end, we have studied Ku70 protein complex and identified several new partners in the nucleus that function in DNA repair and transcriptional regulation. More importantly, we have found that Ku70 played a key role in signal transduction and governed the master switch in apoptotic pathway in the cytoplasm. Currently, we focus our efforts to understand the role of Ku70 in cancer development and other human diseases. Another theme of our research is directed to develop high sensitive structural based proteomic methodologies to elucidate the molecular machine operating inside the cell. We have developed a novel laser induced photochemical protein surface oxidation method to determine protein (or protein complex) solvent accessible surface. We are applying this technology to map sites of protein-protein, protein-DNA/RNA, and protein-drug interactions, and to study their binding kinetics.
In addition, we are developing cutting-edge top-down proteomic technologies based on LC-MS/MS and LTQ Orbitrap Mass Spectrometry. Protein functions are highly regulated by their post translational modifications such as phosphorylation, acetylation and methylation. These modifications are central to cellular signaling pathways and gene regulatory networks, and therefore important for cancer therapy and diagnostics. However, it is difficult to study these modifications using traditional biochemical methods. The top-down proteomic strategy will enable us to characterize and profile global protein post-translational modifications.
SELECTED PUBLICATIONS:
1. Chen L, Sze SK and Yang H, Automated Intensity Descent Method (AID-HRMS) for Efficient Interpretation of Complex High Resolution Mass Spectra of Proteins, Analytical Chemistry, 2006. (In Press).
2. Wong JJY, Pung YF, Sze NSK and Chin KC, HERC5 is a novel interferon-induced HECT-type E3 protein ligase that mediates Type I interferon-induced ISGylation of protein targets, Proceedings of the National Academy of Sciences of the USA, 2006. (Accepted).
3. Guo X, Chan-Park M, Yoon SF, Chun J, Hua L, and Sze NSK, UV Embossed Polymeric Chip for Protein Separation and Identification based on Capillary Isoelectric Focusing and MALDI-TOF-MS, Analytical Chemistry, 78, 3249-3256, 2006.
4. Hua L, Low TY, and Sze NSK, Microwave-assisted Specific Chemical Digestion for Rapid Protein Identification, Proteomics, 6, 586-591, 2006.
5. Aye TT, Low TY, and Sze NSK, Nanosecond Laser Induced Photochemical Oxidation method for Protein Surface Mapping with Mass Spectrometry, Analytical Chemistry, 77(18), 5814-5822, 2005.
6. Mok MLS, Hua Lin, Phua JBC, Wee MKT, and Sze NSK, Capillary Isoelectric Focusing in Pseudo-closed Channel Coupled to Matrix Assisted Laser Desorption/Ionization Mass Spectrometry for Protein Analysis, Analyst, 129, 109-110, 2004.
7. Sze NSK, Ge Y, Oh H, and McLafferty FW, Plasma Electron Capture Dissociation for Large Protein, Analytical Chemistry, 75, 1599-1603, 2003.
8. Ge Y, ElNaggar M, Sze NSK, Hanbin Oh, Begley TP, and McLafferty FW, Top Down Proteomic Analysis of Secreted Proteins from Mycobacterium Tuberculosis by Electron Capture Dissociation Mass Spectrometry, Journal of the American Society of Mass Spectrometry, 14(3), 253-261, 2003.
9. Ge Y, Lwahor BG, ElNaggar M, Sze NSK, Begley TP, and McLafferty FW, Protein Oxidation in Viral Prolyl-4-hydroxylase Characterized by Top Down Tandem Mass Spectrometry, Protein Science, 12, 2320-2326, 2003.
10. Oh H, Breuker K, Sze SK, Ge Y, Carpenter BK, and McLafferty FW, Secondary and tertiary structures of gaseous protein ions characterized by electron capture dissociation mass spectrometry and photofragment spectroscopy. Proceedings Of The National Academy Of Sciences Of The United States Of America, 99, 15863-8, 2002.
11. Sze NSK, Ge Y, Oh H, and McLafferty FW, Top Down Mass Spectrometry of a 29 kDa Protein for Characterization of Any Posttranslational Modification to Within One Residue, Proceedings of the National Academy of Sciences of the USA. 99, 1774-1779, 2002.
PATENTS:
1. Capillary Isoelectic Focusing – MALDI Device for Protein Analysis
2. Polymer Composite for MALDI Analysis of Biomolecules.
Lab Members:
Ran Dong Yuan
Email: rdyuan@ntu.edu.sg
Tel: (65) 6316-2852
Meng Wei
Email: g0402992@nus.edu.sg
Tel: (65) 6316-2852
NARDEV S/O RAMANATHAN
Email: nardev@pmail.ntu.edu.sg
Tel: (65) 6316-2852
Zhao Chen
Email: 149914862@ntu.edu.sg
Tel: (65) 6316-2852
Vacancies:
We are looking for Postdoctoral Fellows, Research Assistants and Graduate Students to join our multi-disciplinary team in the following areas:
1) proteomics technology development: background in physics, chemistry, biology or engineering
2) proteomics bioinformatics/mass spectrometry data analysis: Mathematics, Computer Sciences