Molecular Recognition Laboratorium: Difference between revisions
Victor Tapia (talk | contribs) |
Victor Tapia (talk | contribs) |
||
| (41 intermediate revisions by the same user not shown) | |||
| Line 18: | Line 18: | ||
web charite.de | web charite.de | ||
}} | }} | ||
last change on 01.04.2010 | |||
<br> | <br> | ||
</div><br> | </div><br> | ||
| Line 35: | Line 36: | ||
==Group Members== | ==Group Members== | ||
<div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | <div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | ||
*Annette Hayungs - Secretary {{hide| | *Annette Hayungs - Secretary {{hide| | ||
[[Image:Molrec placeholder.png|130px|right]] | [[Image:Molrec placeholder.png|130px|right]] | ||
| Line 64: | Line 41: | ||
Fax: +49 30 450 524 942 | Fax: +49 30 450 524 942 | ||
E-Mail: annette.hayungs(at)charite.de | E-Mail: annette.hayungs(at)charite.de | ||
| Line 90: | Line 59: | ||
}} | }} | ||
*Judith Müller - Doktorandin {{hide| | *Judith Müller - Doktorandin {{hide| | ||
| Line 131: | Line 85: | ||
}} | }} | ||
*[http://openwetware.org/wiki/User:Victor_Tapia Víctor Tapia] - Doktorand {{hide| | *[http://openwetware.org/wiki/User:Victor_Tapia Víctor Tapia] - Doktorand {{hide| | ||
[[Image:Molrec placeholder.png|130px|right]] | [[Image:Molrec placeholder.png|130px|right]] | ||
ACTUALLY SEARCHING FOR A NEW POSITION | |||
Email: ve.tapia.m@gmail.com | |||
| Line 152: | Line 96: | ||
Fax: +49 30 450 524 942 | Fax: +49 30 450 524 942 | ||
E-Mail: lars.vouilleme(at)charite.de | E-Mail: lars.vouilleme(at)charite.de | ||
| Line 171: | Line 107: | ||
FIGURE {{hide| | FIGURE {{hide| | ||
---- | ---- | ||
[[image:Macro_to_microarrays_of_peptides.png|400px]][[image:Fmoc-spotting.png|400px]] | A_[[image:Macro_to_microarrays_of_peptides.png|400px]] B_[[image:Fmoc-spotting.png|400px]] | ||
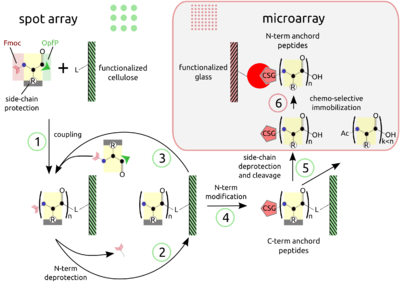
'''PEPTIDE ARRAYS ''' The combination of SPOT peptide synthesis (steps 1 to 4) with appropriate immobilization techniques on glass supports (steps 5 and 6) is wide spread. The SPOT technology provides low-scale but high-throughput synthesis, while immobilization of pre-synthesized peptides offers the benefit of a "chemical" purification step and flexible array design. Additionally, the glass support is compatible with fluorescence detection and offers the possibility to miniaturize binding assays. Beyond economy, the later point is essential for quantitative measurements at the steady-state of binding activity, as has been described [Ekins 1998] and can be proven by the mass-action law. | '''PEPTIDE ARRAYS ''' The combination of SPOT peptide synthesis (figure A, steps 1 to 4) with <br> | ||
appropriate immobilization techniques on glass supports (figure A, steps <br> | |||
5 and 6) is wide spread. The SPOT technology provides low-scale but <br> | |||
high-throughput synthesis, while immobilization of pre-synthesized<br> | |||
peptides offers the benefit of a "chemical" purification step and <br> | |||
flexible array design. Additionally, the glass support is compatible <br> | |||
with fluorescence detection (see figure B, adapted from the web) and <br> | |||
offers the possibility to miniaturize binding assays. Beyond economy, <br> | |||
the later point is essential for quantitative measurements at the <br> | |||
steady-state of binding activity, as has been described [Ekins 1998] and <br> | |||
can be proven by the mass-action law. <br> | |||
---- | ---- | ||
<br> | <br> | ||
}} | }} | ||
<div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | <div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | ||
The basic point of this technology is the simultaneous display of a systematic collection of peptides on a planar support, on which numerous bimolecular interaction assays can be carried out under homogeneous conditions. | The basic point of this technology is the simultaneous display of a <br> | ||
systematic collection of peptides on a planar support, on which numerous <br> | |||
bimolecular interaction assays can be carried out under homogeneous <br> | |||
conditions. <br> | |||
MORE {{hide| | |||
---- | |||
PEPTIDE ARRAYS IN THE ADVANCEMENT OF PEPTIDE SYNTHESIS<br> | |||
<br> | |||
* The development of solid-phase peptide synthesis (SPPS) by Bruce Merrifield [Gutte and Merrifield, 1969; Merrifield, 1965] and adaptions of this procedure [Fields and Noble, 1990] set the chemical ground for innovative technologies to follow. <br> | |||
* The development of the “Pin” method by H. Geysen [Geysen, et al., 1984] introduces the array format to peptide synthesis. <br> | |||
* Definitive establishment of peptide arrays came along with the development of the SPOT synthesis by Roland Frank [Frank, 1992; Frank, 2002] which simplified chemical synthesis of peptide arrays to the addressable deposition of reagents on a cellulose sheet. <br> | |||
<br> | |||
Modern peptide synthesis approaches and <br> | |||
molecular biology make peptides accessible in a high degree of <br> | |||
structural diversity. The two greatest drawbacks of synthetic peptide <br> | |||
arrays are peptide length, with a quality threshold between 30 and 50 <br> | |||
amino-acids, as well as the restriction to linear motives, since the <br> | |||
mimicry of nonlinear motives with linear peptide constructs is still <br> | |||
under development [Goede, et al., 2005]. <br> | |||
<br> | |||
PEPTIDE ARRAYS IN THE ADVANCEMENT OF BINDING ASSAY SYSTEMS <br> | |||
<br> | |||
Since the 90s a major aspect of development to achieve the required <br> | |||
sensitivities to analyse biological samples has been the miniaturization <br> | |||
of analytical devices [Ekins, 1998]. It is important to note that <br> | |||
miniaturization is not only a matter of high-throughput and economy. <br> | |||
Miniaturization is an essential factor that should provide saturation of <br> | |||
binding sites under low analyte concentrations without significantly <br> | |||
altering its bulk (or ambient) concentration upon capturing [Ekins, et <br> | |||
al., 1990; Ekins, 1989; Joos, et al., 2002; Templin, et al., 2002]. | |||
* In this sense, the first application of a peptide microarray device in 1991, anticipating even the application of cDNA arrays, achieved already the impressive feature density of about 1024 peptides in 1.6 cm2 by means of in situ light-directed parallel synthesis [Fodor, et al., 1991]. | |||
Several methods available to generate peptide arrays on planar solid surfaces offer a range between... | |||
* 16 peptides per cm2, in the case of SPOT macroarrays [Reimer, et al., 2002; Schutkowski, et al., 2004], | |||
* to 2000-4000 peptides in 1.5 cm2, in the case of microarrays generated by digital photolithography [El Khoury, et al., 2007; Gao, et al., 2004; Pellois, et al., 2000; Pellois, et al., 2002]. | |||
<br> | |||
SUPPORT MATERIALS <br> | |||
<br> | |||
In order to | |||
support synthesis, planar materials have to fulfil several requirements <br> | |||
including stability towards solvent and reagent deposition. The <br> | |||
functional groups on the surface must also be biochemically accessible <br> | |||
for chemical derivatisation. Furthermore, upon solid-phase binding <br> | |||
assay, generated peptides must be functionally displayed to allow <br> | |||
molecular recognition with a binding partner in the solution phase. In <br> | |||
particular non-specific interactions should be ruled out. | |||
* Flexible porous supports such as cellulose [Eichler, et al., 1989; Frank and Döring, 1988], cotton [Eichler, et al., 1991; Schmidt and Eichler, 1993] or membranes [Daniels, et al., 1989; Wang and Laursen, 1992; Wenschuh, et al., 2000] are preferentially used for peptide array generation. <br> | |||
* Rigid, non-porous materials such as glass [Falsey, et al., 2001], gold films [Houseman and Mrksich, 2002; Jonsson, et al., 1991; Malmqvist, 1993], or silicon [Fodor, et al., 1991; Pellois, et al., 2002] have also been used for in situ synthesis, but are much more technically demanding. <br> | |||
* On the other side, rigid materials have a number of advantages over porous supports for functional display of molecules. Impermeability and smooth two-dimensionality of the material does not limit diffusion of the binding partner and leads to more accurate kinetics of recognition events. <br> | |||
* Finally the flatness and transparency of glass improve image acquisition and simplifies the use of fluorescence dyes for the read out process. <br> | |||
In some cases, assembled 3D structure on a non-porous surface could be <br> | |||
a fruitful approach. Several techniques for coherent surface modifications <br> | |||
are described over the past twenty years in the literature. For a comparative <br> | |||
overview on this field we refer to articles dedicated to the peptide and protein array <br> | |||
technologies [Angenendt and Glokler, 2004; Angenendt, et al., 2002; <br> | |||
Angenendt, et al., 2003; Seurynck-Servoss, et al., 2008; <br> | |||
Seurynck-Servoss, et al., 2007; Seurynck-Servoss, et al., 2007; Sobek, <br> | |||
et al., 2007; Sobek, et al., 2006; Wenschuh, et al., 2000]. <br> | |||
}} | |||
REFERENCES {{hide| | |||
---- | |||
* Angenendt, P., and Glokler, J. (2004) Evaluation of antibodies and microarray coatings as a prerequisite for the generation of optimized antibody microarrays, Methods Mol Biol 264, 123-34. | |||
* Angenendt, P., Glokler, J., Murphy, D., Lehrach, H., and Cahill, D. J. (2002) Toward optimized antibody microarrays: a comparison of current microarray support materials, Anal Biochem 309, 253-60. | |||
* Angenendt, P., Glokler, J., Sobek, J., Lehrach, H., and Cahill, D. J. (2003) Next generation of protein microarray support materials: evaluation for protein and antibody microarray applications, J Chromatogr A 1009, 97-104. | |||
* Daniels, S. B., Bernatowicz, M. S., Coull, J. M., and Köster, H. (1989) Membranes as solid supports for peptide synthesis., Tetrahedron Lett. 30. | |||
* Eichler, J., Beyermann, M., and Bienert, M. (1989) Application of cellulose paper as support in simultaneous solid phase peptide synthesis., Colect. Czech. Chem. Commun. 54, 1746-52. | |||
* Eichler, J., Bienert, M., Stierandova, A., and Lebl, M. (1991) Evaluation of cotton as a carrier for solid-phase peptide synthesis., Peptide Res. 4, 296-307. | |||
* Ekins, R., Chu, F., and Biggart, E. (1990) Multispot, multianalyte, immunoassay, Ann Biol Clin (Paris) 48, 655-66. | |||
* Ekins, R. P. (1989) Multi-analyte immunoassay, J Pharm Biomed Anal 7, 155-68. | |||
* Ekins, R. P. (1998) Ligand assays: from electrophoresis to miniaturized microarrays, Clin Chem 44, 2015-30. | |||
* El Khoury, G., Laurenceau, E., Dugas, V., Chevolot, Y., Merieux, Y., Duclos, M. C., Souteyrand, E., Rigal, D., Wallach, J., and Cloarec, J. P. (2007) Acid deprotection of covalently immobilized peptide probes on glass slides for peptide microarrays, Conf Proc IEEE Eng Med Biol Soc 2007, 2242-6. | |||
* Falsey, J. R., Renil, M., Park, S., Li, S., and Lam, K. S. (2001) Peptide and small molecule microarray for high throughput cell adhesion and functional assays, Bioconjug Chem 12, 346-53. | |||
* Fields, G. B., and Noble, R. L. (1990) Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids, Int J Pept Protein Res 35, 161-214. | |||
* Fodor, S. P., Read, J. L., Pirrung, M. C., Stryer, L., Lu, A. T., and Solas, D. (1991) Light-directed, spatially addressable parallel chemical synthesis, Science 251, 767-73. | |||
* Frank, R. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support, Tetrahedron, 9217-32. | |||
* Frank, R. (2002) The SPOT-synthesis technique: Synthetic peptide arrays on membrane supports--principles and applications, J. Immunol. Methods 267, 13-26. | |||
* Frank, R., and Döring, R. (1988) Simultaneous multiple peptide synthesis under continuous flow conditions on cellulose paper disks as segmental solid supports, Tetrahedron 44, 6031-40. | |||
* Gao, X., Pellois, J. P., Na, Y., Kim, Y., Gulari, E., and Zhou, X. (2004) High density peptide microarrays. In situ synthesis and applications, Mol Divers 8, 177-87. | |||
*Geysen, H. M., Meloen, R. H., and Barteling, S. J. (1984) Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid, Proc Natl Acad Sci U S A 81, 3998-4002. | |||
*Goede, A., Jaeger, I. S., and Preissner, R. (2005) SUPERFICIAL--surface mapping of proteins via structure-based peptide library design, BMC Bioinformatics 6, 223. | |||
*Gutte, B., and Merrifield, R. B. (1969) The total synthesis of an enzyme with ribonuclease A activity, J Am Chem Soc 91, 501-2. | |||
*Houseman, B. T., and Mrksich, M. (2002) Towards quantitative assays with peptide chips: a surface engineering approach, Trends Biotechnol 20, 279-81. | |||
*Jonsson, U., Fagerstam, L., Ivarsson, B., Johnsson, B., Karlsson, R., Lundh, K., Lofas, S., Persson, B., Roos, H., Ronnberg, I., and et al. (1991) Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology, Biotechniques 11, 620-7. | |||
*Joos, T. O., Stoll, D., and Templin, M. F. (2002) Miniaturised multiplexed immunoassays, Curr Opin Chem Biol 6, 76-80. | |||
*Malmqvist, M. (1993) Biospecific interaction analysis using biosensor technology, Nature 361, 186-7. | |||
*Merrifield, R. B. (1965) Automated synthesis of peptides, Science 150, 178-85. | |||
*Pellois, J. P., Wang, W., and Gao, X. (2000) Peptide synthesis based on t-Boc chemistry and solution photogenerated acids, J Comb Chem 2, 355-60. | |||
*Pellois, J. P., Zhou, X., Srivannavit, O., Zhou, T., Gulari, E., and Gao, X. (2002) Individually addressable parallel peptide synthesis on microchips, Nat Biotechnol 20, 922-6. | |||
*Reimer, U., Reineke, U., and Schneider-Mergener, J. (2002) Peptide arrays: from macro to micro, Curr Opin Biotechnol 13, 315-20. | |||
*Schmidt, M., and Eichler, J. (1993) Multiple peptide synthesis using cellulose-based carriers: Synthesis of substance P - diastereoisomers and their histamine-releasing activity., Bioorg. Med. Chem. Lett. 3, 441-46. | |||
*Schutkowski, M., Reimer, U., Panse, S., Dong, L., Lizcano, J. M., Alessi, D. R., and Schneider-Mergener, J. (2004) High-content peptide microarrays for deciphering kinase specificity and biology, Angew Chem Int Ed Engl 43, 2671-4. | |||
*Seurynck-Servoss, S. L., Baird, C. L., Miller, K. D., Pefaur, N. B., Gonzalez, R. M., Apiyo, D. O., Engelmann, H. E., Srivastava, S., Kagan, J., Rodland, K. D., and Zangar, R. C. (2008) Immobilization strategies for single-chain antibody microarrays, Proteomics 8, 2199-210. | |||
*Seurynck-Servoss, S. L., Baird, C. L., Rodland, K. D., and Zangar, R. C. (2007) Surface chemistries for antibody microarrays, Front Biosci 12, 3956-64. | |||
*Seurynck-Servoss, S. L., White, A. M., Baird, C. L., Rodland, K. D., and Zangar, R. C. (2007) Evaluation of surface chemistries for antibody microarrays, Anal Biochem 371, 105-15. | |||
*Sobek, J., Aquino, C., and Schlapbach, R. (2007) Quality considerations and selection of surface chemistry for glass-based DNA, peptide, antibody, carbohydrate, and small molecule microarrays, Methods Mol Biol 382, 17-31. | |||
*Sobek, J., Bartscherer, K., Jacob, A., Hoheisel, J. D., and Angenendt, P. (2006) Microarray technology as a universal tool for high-throughput analysis of biological systems, Comb Chem High Throughput Screen 9, 365-80. | |||
*Templin, M. F., Stoll, D., Schrenk, M., Traub, P. C., Vohringer, C. F., and Joos, T. O. (2002) Protein microarray technology, Drug Discov Today 7, 815-22. | |||
*Wang, Z., and Laursen, R. A. (1992) Multiple peptide synthesis on polypropylene membranes for rapid screening of bioactive peptides., Pep. Res. 5, 275-80. | |||
*Wenschuh, H., Volkmer-Engert, R., Schmidt, M., Schulz, M., Schneider-Mergener, J., and Reineke, U. (2000) Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides, Biopolymers 55, 188-206. | |||
}} | |||
</div> | |||
<br> | |||
===Messsystemanalyse von Peptidarray Techniken=== | |||
by ''Victor Tapia'' <br> | |||
<div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | |||
Mit dem Ziel die Affinitätsvorhersagbarkeit von Peptidarray-orientierten Bindunsstudien zu analyzieren, wurde die CB4 1 Epitope Bibliothek definiert, welche auf ein Repertoire von 26 verschiedene Liganden für das CB4-1 Antikörper besteht. Es handelt sich hierbei haupsächlich um Sequenz- und Mimika¬varianten des natürlichen Epitopes von CB4-1 (? p24 von HIV 1). Die Eignung dieser Proben für das oben erklärtes Ziel beruhrt auf das vorhandene Affinitätsprofil, das jedes Peptid mit einzeln gemessenen KD Werte assoziiert. <br> | |||
<br> | |||
Die einzelne KD Messungen wurden teilweise aus der Literatur aussortiert [Volkmer-Engert 1995; Kramer 1997; Kramer 1998; Hoffmuller 2000; Reineke 2002] und teilweise mit eigenen SPR Messungen belegt [Weiser 2005].<br> | |||
<br> | |||
Das Peptid Repertoire der Bibliothek umfasst ein Affinitätsbereich mit KD-Werten zwischen 1 mM und 1 nM. Diese Bibliothek wird sowohl zur Analytbestimmung wie auch zur Affinitätsvorhersage eingesetzt, um die Bediengungen eines Assays zu determinieren, unter welcher eine wertgenaue Vorhersage möglich ist. <br> | |||
<br> | |||
Hierbei handelt es sich um ein Bindungsstudie in Mikroarray-Format [Reimer 2002; Tapia 2009]. Die Wert¬genauig¬keit der Ansatz wird anhand der Übereinstimmung mit Referenz Affinitätswerten (Sollwerte) aus den einzelne SPR-bezogene Bindunsstudien. Der Einsatz <br> | |||
dieser Referenzmesswerte als „goldener Standard“ ist begründet durch drei wessentliche unterschiede zu den Mikro- und Makroarray Formate der HTS Bindungsstudien: (1) die Peptid Proben sind ausführlich präpariert worden (HPLC, MS), (2) das Analyt wurde durch die Messzelle des Sensorchips durchflossen und (3) die Auswertung geschieht anhand von Sensogrammen, die eine echtzeit Beobachtung der Gleichgewichtzustandes ermöglicht (siehe Abbildung 1 für ein Überblick).<br> | |||
<br> | |||
'''MORE TEXT 1''' {{hide| | |||
---- | |||
Die 26 Epitopvarianten der CB4-1 Epitopebibliothek wurden als Amino-oxyacetylierte Peptid-Proben via Spot Technik synthetiziert. Nach der Abspaltung der Schutzgruppen der Seitenkette wurden die Proben auf eine Mikrotitterplatte transferiert. Anschlißend, wurden die Rohprodukten der Proben auf Aldehyd-funktionalizierte Glass-Trägern mit einem piezoelektrischen Printer immobiliziert. Dadurch wurden die Peptide von N- (gebunden) zu C-Term (frei) präzentiert. Die Arrayanordnung ist in Abbildung 2 dargestellt. Für die vier Wiederholungen der dort gezeigten Bindunsstudie, stammten die funktionalizierte Plattformen aus einem Produktions-„batch“ des Herstellers (Genetix Ltd., UK) und ebenso wurden in einem „batch“ darauf die Peptidproben immobiliziert und mit dem CB4 1 Analyt inkubiert. Ein Minimum an technische Quellen für Variation in den Messungen wurde in dieser Weise angestrebt. | |||
Abbildung 2. | |||
A.- Die vier Spalten zeigen die Negativ-Verarbeitung | |||
der Bildaufzeichnungen von vier Wiederholungen eines | |||
Mikroarray Bindungsstudie der CB4-1 Antikörper auf 20 | |||
Replikas eines 4?8 Arrays (Panel C). Die 20 Arrays jedes | |||
Plattforms sind vertikal in fünf Felder von 4 Arrayreplikas | |||
angeordnet. | |||
B.- Vergrößerung des viereckigen Rahmen im Panel A, d.h. | |||
ein Replikafeld. Die aufgezeichneten Signalintensitäten | |||
sind, soweit unspezifische Wechselwirkungen des | |||
Detektionsantikörpers ausgeschlossen werden konnten, von | |||
den Mengen an Analyt abhängig, die auf jeden Peptidspot | |||
eingefangen wurden. | |||
C.- Die Positionen des 4?8 Arrays wurden mit den | |||
Peptidsequenzen der Subbibliothek besetzt. Die angegebene | |||
Zahlen auf jeden Spot entsprechen die interne | |||
ID-Erkennungszeichen aus Tabelle 1. Die Sequenzspezifität | |||
des Immunoanalyts (?-p24 HIV-1, CB4-1) kann aus der | |||
Zusammenschluß beider Informationen, d.h. positionsabhängige | |||
Signalintensitäten und adressierbare Sequenzliste, abgeleitet | |||
werden. Die Abbildung zeigt insgesamt 80 Wiederholungen von | |||
Proben 1, 3-17, und 19-26 sowie 320 Wiederholungen von | |||
Proben 2 (schwach Binder) und 18 (stark Binder). | |||
<br> | |||
<nowiki>Die Qualität der Fluoreszenzmessungen wurde in zwei Hinsichten | |||
untersucht: Präzision und Wertgenauigkeit. Die Präzision der Technik | |||
wurde mittels eine beschreibende Untersuchung der Variation in der | |||
Messung charakterisiert. Dabei wurde die Streuung von Signalintensitäten | |||
aus Replikaspots, die innerhalb eines Plattforms verteilt waren, durch | |||
das Variations¬koeffizient (engl. „coefficient of variation“, COV= | |||
Standard Abweichung/Mittelwert) erfasst. Im Durchschnitt, ergibt das COV | |||
der vier Plattformen ein Wert von 0,31 ± 0,177 (Abbildung 3, Panel A). | |||
Dieser Wert entspricht die Streuung der Signalintensität | |||
(Wiederholpräzision, engl. repeatability), die bei äquivalenten | |||
Messungen innerhalb eines Plattforms erwartet wird. Wiederum, der | |||
paarweise Vergleich der kompleten Datensätze jedes Plattforms mit | |||
einander durch lineare Korrelationsanalyse ist in der Lage die | |||
Nachvollziehbarkeit (Vergleichspräzision, engl. reproducibility) der | |||
gemessene Signalintensitäten zu beschreiben. Im Durchnitt, das | |||
Determinationskoeffizient der 6 nicht-redundanten paarweisen Vergleiche | |||
ergibt 0,98 ± 0,013 (Abbildung 3, Panel B). Dieser Wert beschreibt eine | |||
technische Fähigkeit, indem von 100 Vergleiche 98 nicht unterscheidbare | |||
Messungen zu erwarten sind. Eine Erweiterung dieser Untersuchung auf 3 | |||
weitere unterschiedliche Plattform Produkte und 2 Liefermengen (engl. | |||
batches) wurde in [Tapia 2007] veroffentlicht. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Abbildung 3. Panel A: Wiederholpräzision des Mikroarray-Messsystems. Die Fähigkeit, ein kohärentes Mittelwert von Replikas innerhalb einer Messung zu wiederspiegeln, ist für vier Plattforms (MS1-4), die wie im Abbildung 2 mit Peptid Proben belegt wurden, beschrieben. Der globale Durchschnitt der COV Werte (gestrichelte Linie) ist 0,31 ± 0,177. Panel B: Vergleichspräzision des Mikroarray-Mess¬systems. Die Fähigkeit, kohärente Messergebnisse mit unabhängigen Messungen zu erzielen, ist durch die Bestimmtheitsmaß (R2) von paarweisen Vergleichen der Fluoreszenz (engl. mean SI) beschrieben. Im Durchschnitt R2=0,98 ± 0,013. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Die Wertgenauigkeit (oder Richtigkeit, engl. accuracy) der Signalmessungen wird mittels einer analytischen Untersuchung zweierlei Effekten auf die von Arrayproben eingefangene Analytmenge beschrieben: erstens, der Effekt von variierenden Affinitäts¬werte (Analyt-bestimmung) und, zweitens, der Effekt von variierenden Analyt¬konzentrationen (Affinitäts-bestimmung). Hierbei unterscheidet man zwischen Soll- und Istwerte (engl. „nominal“ und „actual values“, entsprechend). Bei der Analytbestimmung ist die experimentell justierbare Variable durch unabhängig bestimmte Affinitäten der Peptidproben repräsentiert. Für zwecken der Analyse wird der Einfluß dieser Variable auf die eingefangene Analytmenge (und dadurch auf die gemessene Signalintensitäten) bei eine Serie von unterschiedlich eingesetzten Analytkonzentrationen untersucht. Die Sollwerte der Analytbestimmung sind somit durch die fünf verwendete molare Konzentrationen (cAN={3,34 µM; 667 nM; 66,7 nM; 6,67 nM und 667 pM}) vertretten. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Tabelle 3. Vergleich zwischen beider Ansätze der Vorhersage. | |||
Ansatz experimentell justierbare Variable Istwert Sollwert Model | |||
Analytbestimmung 26 pKD Werte cA cAN ?=?(pKD) | |||
Affinitätsbetimmung 5 cA Werte EC50 pKDN ?=?(cA) | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Dagegen, ist bei der Affinitätsbestimmung die experimentell justierbare Variable durch eine Reihe von bestimmten Analytkonzentrationen repesentiert. Für zwecken der Analyse wird der Einfluß dieser Variable bei eine Serie von unabhängig gemessenen Affinitäten untersucht. Die Sollwerte der Affinitätsbestimmung sind somit durch die 26 verwendete Affinitäten (pKDN aus Tabelle 1) vertretten. | |||
Die Istwerte (engl. actual values) werden durch die geschätzte Werte, die mit der jeweiligen Lösungsansätzte der selber Regressionsmodel erzielt wurden, representiert. Die Analytbestimmung ergibt sich aus der Formulierung der Regressionsmodel als ?=?(pKD) mit dem Systemkonstante cA. Für jeder Inkubation werden die cA als Istwerte geerntet. Durch eine Reformulierung des Regressionsmodels als ?=?(cA) wird bei der Affinitätsbestimmung das Systemkonstanten KD jeder Peptidprobe als Istwert geerntet. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Abbildung 4. Wertgenauigkeit der Analytbestimmung. Hier handelt es sich um ein Gleichgewichtsanalyse der Effekt von variierten Affinitätswerten (einzelne SPR pKD Werte) auf die Analytmenge, die von den Arrayproben eingefangen werden. Panel A und B zeigen die Streuung der SI Messdaten und die Regressionskurven, die auf unabhängige Inkubationen der Arrayproben mit einer 3,34 µM bzw. einer ca. 0,67 nM Analytkonzentration (cAN Sollwert, engl. nominal value), entsprechend, zurückzuführen sind. Panel C fasst die Regressionskurven für alle angewandte Analyt¬konzentrationen (cAN= {3,3 µM; 670 nM; 67 nM; 6,7 nM und 0,67 nM}) zusammen. Die dazu gehörige Parameterwerte (cA Istwerte, engl. actual cA) sind im Panel D mit cAN. Zur Diskussion der Kurvenverläufen in Panel D ist außerdem dem geschätzten Wert der Peptidkonzentration (cP Istwert, engl. cP actual value) vergleichend dargestellt. | |||
</nowiki> | |||
<br> | <br> | ||
</div><br> | <nowiki> | ||
Abbildung 4 fasst die Ergebnisse der Analytbestimmung zusammen. Panel A und B beweisen einen mit der Messdaten koherente Verlauf der Regressionskurven für zwei der angewandeten Analytkonzentrationen, 3,34 µM und 0,67 nM entsprechend. Ebenso in Panel C ersichtlich ist ein mit den angewandten Analytkonzentrationen koherenten Verrückung der Regressionskurven. Auf dem ersten Blick scheint die niedrigste Konzentration (0,67 nM) dieser Beobachtung zu wiedersprechen. Dafür werden in Panel D die Werte der Analytbestimmung mit dem geschätzten Wert der Peptidkonzentration verglichen. Offensichtlich, ist im Messsystem eine 0,67 nM Konzentration des Analyts niedriger als die Peptidkonzentration. Dieser Zustand führt zu einer im Gleichgewicht unvollständigen Besetzung der Bindungstellen. Einer Erhöhung der Analytkonzentration muss zuerst die vollständige Besetzung der Bindungstellen bewirken bevor die Regressionskurven höhere Analytkonzentrationen entsprechend nach links im Panel C verrücken können. Dieser Möglichkeit soll während des fitting-Verfahrens unbedingt beachtet werden. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Die Wertgenauigkeit der Analytbestimmung ist durch eine paarweise lineare Korrelation der Ist- und Sollwerte der Analytkonzentrationen mit einem Bestimmtheitsmaß R2=0.98 charakteriziert. Allerdings, eine systematische Messabweichung zu niedrigen Istwerte bei Sollwerte > 67 nM und zu höheren Istwerte bei Sollwerte < 67 nM ist ersichtlich. Es ist aus dieser Analyse nicht zu entnehmen, ob eine Erwieterung der angewendten Anaylt-konzentrationen mit extremeren Werten zu einer Bestätigung oder Ausgleich der systematischen Abweichung führen kann. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Von unmittelbare Interesse für die gegenwärtige Arbeit is die Bestimmung von Affinitätswerte. Solches Ziel gelingt durch die Untersuchung der Abhängigkeit der eingefangenen Analytmenge von der Analytkonzentration (justierbare Variable). Hierbei handelt es sich um die statistische Schätzung (oder, etwas optimitischer, die Vorhersage) der Systemkonstanten (insbesondere Istwerte, negativen dekadischen Logarithmus der Dissoziations¬konstante KD in mol*L-1). Die Genauigkeit der Vorhersage lässt sich durch lineare Korrelation mit dem Affinitätssollwerten (negativen dekadischen Logarithmus aus unabhängigen einzelne KDN Messungen in mol*L-1) bestimmen. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Abbildung 5 fasst die Ergebnisse der Affinitätsbestimmung in Analogie zu Abbildung 4 zusammen. Panel A und B zeigen die kohärenz der Kurvenverläufe in Bezug auf die Verteilung der Messdaten mit Bestimheitsmaßen von R2=0,98 (schwacher Binder, pKDN= 3,52) und 0,95 (starker Binder, pKDN= 8,22), entsprechend. Um die Abhängigkeit der SI von variirenden Analytkonzentrationen darzustellen, wurde die Anordnung der Datensätze im Vergleich zu Abbildung 4 transponiert. <br> | |||
Dadurch findet in Panel A bis C ein Kurvenverlauf von einem SI Minimum zu einem Maximum in Umgekehrte Richtung statt. Panel C zeigt die durch die sinkende Affinitätsistwerte definierte links-Verrückung der Kurvenverläufen. Die Regressionskurven erreichen asymptotisch ein global definiertes SI Maximum. | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Im Panel D von Abbildung 5 ist eine kaum lineare Vergleich der Messdaten ersichtlich (R2?0,70). Zwei Aspekte verdienen besondere Kritik und werden die Versuchsplannung der PQBP-1 Bindungsstudien entscheidend Prägen. Erstens, Sollwerte zwischen 3,5 und 6,2 werden von Istwerte begegnet, die zwischen 4,2 und 5.8 einengent bestimmt wurden. Zweitens der schmalen dynamischen Bereich bei Sollwerten von 6,2 bis etwa 7,3 wird von kaum korrelierenden Istwerten begegnet (R2<0,50). | |||
</nowiki> | |||
<br> | |||
<nowiki> | |||
Abbildung 5. Wertgenauigkeit der Affinitätsvorhersage. Hier handelt es sich um ein Gleichgewichtsanalyse der Effekt von variierten Analytkonzentrationen auf die Analytmenge die von den Arrayproben eingefangen werden. Panel A und B zeigen die Streuung der SI Messdaten und die Regressionskurve für die Inkubation eines belegten Plattforms mit einer etwa 3 µM und einer etwa 0,7 nM Analytkonzentration (cA Sollwert, engl. nominal value), entsprechend. Panel C fasst die Regressionskurven für alle angewandte Analytkonzentrationen zusammen. Die dazu gehörige Parameterwerte (cA Istwert, engl. actual value) sind im Panel D mit dem erwarteten cA Sollwert und dem geschätzten Wert der Peptidkonzentration (cP Istwert, engl. cP actual value) vergleichend dargestellt. | |||
</nowiki> | |||
}} | |||
</div> | |||
<br> | |||
===Structural Modularity in Protein-Protein Recognition=== | ===Structural Modularity in Protein-Protein Recognition=== | ||
by ''Victor Tapia'' <br> | by ''Victor Tapia'' <br> | ||
FIGURE {{hide| | FIGURE {{hide| | ||
---- | ---- | ||
[[ | [[Image:Seet 2006 - Src PTM switch.tif|400px]] | ||
image was adapted from Seet et al 2006 | |||
---- | ---- | ||
}} | }} | ||
====Protein Interaction Domains==== | ====Protein Interaction Domains==== | ||
<div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | <div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | ||
The organisation of living systems is a complex network of molecular interactions. Proteins are a central component of the network as they may bind to other proteins as well as to phospholipids, nucleic acids and small molecules to interconnect the diverse physiological functions of the cell. In the background of these observations, the existence of a molecular recognition code for cellular organisation is very suggestive. <br> | The organisation of living systems is a complex network of molecular interactions. Proteins are a central component of the network as they may bind to other proteins as well as to phospholipids, nucleic acids and small molecules to interconnect the diverse physiological functions of the cell. In the background of these observations, the existence of a molecular recognition code for cellular organisation is very suggestive. <br> | ||
'''MORE TEXT''' {{hide| | '''MORE TEXT''' {{hide| | ||
---- | ---- | ||
Structural analysis of functional protein complexes suggests at least two classes of protein-protein interactions. In the first class, the so called surface/surface interactions, complementary rigid surfaces link the interacting partners. Under these circumstances, the residues involved in each interacting surface only come together upon protein folding. The second class consists on asymmetric or surface/string interactions, where a protein domain (folding module of moderate size that may resemble a pocket on the protein’s surface) docks a short lineal peptide motive (a peptide ligand) on the partner protein. While surface/surface interactions cannot be inferred because the primary structure is not linearly involved, the binding determinants of a protein interaction domain (PID) may be mapped to short peptides matching the sequence of the ligand peptide. The importance of these recognition domains in the formation of protein complexes by binding to short lineal peptides was firstly demonstrated in the late 1980s and early 1990s [Sadowski 1986 Dec; Mayer 1993; Ren 1993].<br> | |||
Structural analysis of functional protein complexes suggests at least two classes of protein- | |||
<br> | <br> | ||
In | In a pioneering work on the kinase function and transforming activity of the Fujinami Sarcoma Virus, Sadowski et al.[Sadowski 1986 Dec] discovered “a unique domain… (which) is absent from kinases that span the plasma membrane” and concluded that “the presence of this noncatalytic domain in all known cytoplasmic tyrosine kinases of higher and lower eukaryotes argues for an important biological function... the noncatalytic domain may direct specific interactions of the enzymatic region with cellular components that regulate or mediate tyrosine kinase function”. These regions were called Src homology 2 (SH2) and 3 (SH3), the name SH1 being reserved to the catalytic region. Since then, the gained knowledge on SH2/3 domain function has been a paradigm in our understanding of PID biochemistry, e.g. PIDs as non-catalytic binding sites and with modular structure.<br> | ||
<br> | <br> | ||
In this era of extensive genome sequencing, many PIDs have been discovered. The interaction partners — and, therefore, the functions of many proteins ― may be determined by identifying the critical binding sites for family members through evolutionary tracing [Lichtarge 1996 Mar 29] or mapping protein-protein interactions using functional protein arrays [Phizicky 2003 Mar 13]. Many of the PIDs in proteins can be grouped into families that show clear evidence of their evolution from a common ancestor, and genome sequences from Saccharomyces cerevisiae to Homo sapiens reveal large numbers of proteins that contain one or more common domain. | |||
}} | }} | ||
</div><br> | </div> | ||
<br> | |||
==== | ====Modularity of WW PIDs==== | ||
<div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | <div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | ||
The WW domain is a well-characterized protein module that mediates specific protein-protein interactions with ligands that contain short proline-rich motifs [Bork 1994; Chen 1995; Sudol 2000]. The domain is composed of 38 amino acids, it has two signature tryptophan (W) residues, and it is one of the smallest among modular protein domains. The signature tryptophan positions are spaced 20–22 amino acids apart and have important structural role. The structure is a three beta-strand meander that forms a shallow binding pocket for proline-rich ligands (reviewed in[Sudol 2005]). In general, the folding of WW domains does not require cognate ligands or co-factors. <br> | |||
'''MORE TEXT''' {{hide| | '''MORE TEXT''' {{hide| | ||
---- | ---- | ||
There are two aspects which allow a protein motiv to be recognized as module. Firstly, '''functional modularity''': a domains ability to function independent of context, allowing transfer of function between diverse molecular systems. In the case of a WW domain, it functionally resembles the Src homology domain 3 in its recognition of proline-rich or proline-containing ligands. Based on this ligand predilection, two major and two minor groups are classified:<br> | |||
<br> | <br> | ||
Group I core consensus PPxY | |||
Group II PPLP motif | |||
Group III poly-P motifs flanked by R or K | |||
Group IV short seq. with pS or pT followed by P <br> | |||
<br> | <br> | ||
The | The involved affinity of interactions in terms of KD for polyproline rich ligands (Groups I to III) range between µM to nM values. For pSP- or pTP-containing ligands, binding strengths are measured in the low µM range. <br> | ||
<br> | <br> | ||
Secondly, '''structural modularity''': the ability of a protein motiv to be physically isolated from its protein context and yet fold into the characteristic conformation (structural autonomy). Although several WW domain structures are annotated in the Protein Database (PDB), some can only be shown to fold stabily upon ligand binding. It is a matter of controversy, if ligand binding induces the fold or if it selects a stable bounded conformation from many dynamically interchanging free conformations. This is analogous to the question whether ligand binding or folding firstly occurs. <br> | |||
}} | }} | ||
</div><br> | </div> | ||
<br> | |||
====From Promiscuous Recognition Events to Mutually Exclusive Cellular Responses==== | ====From Promiscuous Recognition Events to Mutually Exclusive Cellular Responses==== | ||
<div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | <div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | ||
The elucidation of functional pathways of signal transduction, biochemical function or gene regulation, is firstly addressed in proteomics by deriving interaction networks depicting ideally all interactions in the cell. Several attempts have been done in this direction on different model organisms and with varied methods, including co-purification by affinity chromatography | The elucidation of functional pathways of signal transduction, biochemical function or gene regulation, is firstly addressed in proteomics by deriving interaction networks depicting ideally all interactions in the cell. Several attempts have been done in this direction on different model organisms and with varied methods, including co-purification by affinity chromatography [Gavin 2002; Ho 2002; Bouwmeester 2004], yeast two-hybrid [Uetz 2000; Ito 2001], phage display, peptide array technologies [Landgraf 2004], etc. A comparison of datasets derived by individual methods demonstrates that different methods have different potential. <br> | ||
'''MORE TEXT''' {{hide| | '''MORE TEXT''' {{hide| | ||
---- | ---- | ||
For example, affinity chromatographic approaches are biased to tight interactions such as those involving extensive complementary surfaces, while interactions in which one of the two partners contains at least one PID are more frequent in the two-hybrid database. The higher sensitivity of the so called synthetic approaches (yeast two-hybrid, phage display and peptide array technologies) make them better suited for detecting PID-mediated interactions since their peptide affinity in terms of KD falls in the 10 – 100 µM range (high Kd --> low affinity). However, this advantage is counterbalanced by a low specificity, especially of the yeast two-hybrid approach [Phizicky 2003]. <br> | |||
In order to correct this deficiency a double check-up of the information fed into the interaction databases is recommended. This can be achieved by deriving two interaction networks through orthogonal (fundamentally different) synthetic methods and then considering only the intersection between the two datasets | <br> | ||
In order to correct this deficiency a double check-up of the information fed into the interaction databases is recommended. This can be achieved by deriving two interaction networks through orthogonal (fundamentally different) synthetic methods and then considering only the intersection between the two datasets [Tong 2002; Castagnoli 2004; Landgraf 2004]. False positive reports are thus reduced if the causes for measurement error are different in each method.<br> | |||
<br> | |||
The strength of this combined approach to deliver physiologically relevant interactions has been proven for a phage display/yeast two-hybrid intersected dataset [Tong 2002]. A notable conclusion of this approach is that the intersected dataset of proteins that are able to interact with a given PID is larger than expected when cellular events are viewed as a precise wiring of the proteins in the cell. Although a set of these biochemically potential binders may have no physiological relevance due to expression at different times or tissues, in vitro disrupted structures, etc., the paradox of promiscuous recognition and mutually exclusive responses seems to be inherent to PID mediated interactions: the work of Landgraf et al. [Landgraf 2004] supports the observation that a large fraction of natural peptides with the biochemical potential to bind to any given SH3 domain is actually used in vivo to mediate the formation of a complex.<br> | |||
<br> | |||
An additional difficulty to derive functional interaction pathways is that the difference in affinity between ‘specific’ and ‘non-specific’ interactions has been shown to be less than two orders of magnitude in the case of SH2 and its peptide ligands [Songyang 2004]. Even when granted that the recognition specificity of intact proteins by SH3 domains is greater than for SH3-peptide recognition, affinity is not raised above one order of magnitude [Lee 1995 Oct 16; Arold 1998 Oct 20]. Moreover, the ability of a point-mutant Src SH2 domain to effectively substitute for the SH2 domain of the Sem-5 protein in activation of the Ras pathway in vivo emphasises that the specificity of Sh2-mediated interactions is not great [Marengere 1994]. Consider the later statements under the light of the fact that the affinity of the protein OppA for its ligands is in the range of two orders of magnitude (Oppa is involved in the mopping of peptides in the bacterial periplasm exhibiting no sequence specificity). Tu put it all into a nutshell: the described facts lead to a view of large and promiscuous SH-mediated interaction networks.<br> | |||
<br> | <br> | ||
Since it is possible to generate mutant SH3 domains that have up to 40-fold higher affinity than their wild-types [Hiipakka 1999] , the potential of these domains as research tools and as source of lead compounds for pharmaceutical development can not be overseen. Furthermore, a question cannot be overheard in our minds: which is the functional advantage of maintaining relative low affinity and selectivity for PID-mediated interactions, instead of optimizing the potential affinity of PIDs? And further: how can PID-dependent interaction pathways achieve precise cellular responses?<br> | |||
<br> | <br> | ||
A comfortable view is that sufficient effective selectivity can be brought by compartmentalization, additive effects of multiple separate interactions, cooperative assembly of multiprotein complexes, etc. and that these effects can sustain linear functional pathways. An interesting insight into this mater has been advanced by Zarrinpar, A. et al. In their work [Zarrinpar 2003 Dec 11] the authors find out that while metazoan SH3 domains may rescue the functionality of mutated Sho1-SH3 of the yeast, in the set of yeast-own SH3 domains this promiscuity is forbidden. They thus conclude and confirm that, on the background of diverging SH3 domains, negative selection has drifted the ligand sequences to non-overlapping areas of the particular SH3 binding regions on the sequence space.<br> | |||
<br> | <br> | ||
Alternatively, a divergence process of the shape of the binding region may be ‘guided’ by positive selection to avoid overlapping and, thus, promiscuitive interactions. Nevertheless, the picture of linear functional pathways is being revolutionized by a more probabilistic view of a dynamical equilibrium between multiple interactions, in which “the central organizing principle is a vast and ever-shifting web of interactions, from which output is gauged by global changes in complex binding equilibria”[Mayer 2001 Apr]. Cellular systems are explained here as transformational systems in which a redundant or degenerate input is transformed into a meaningful pattern. Irun Cohen’s ‘emergent specificity’, as postulated to synthesize the clonal selection theory and our knowledge on the promiscuity of immunoreceptors[Cohen 2001; Cohen 2001; Parnes 2004], aims at stitching this fissure with nonlinearity. Emergent properties are those behaviours manifested by a system when it operates as a whole, with no straightforward reducibility to the particulate components taking part in the process. | |||
<br><br><br> | |||
}} | |||
'''REFERENCES''' {{hide| | |||
---- | |||
<br><br> | |||
Sadowski, I, JC Stone & T Pawson (1986 Dec). A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol 6(12): 4396.<br> | |||
<br> | |||
Mayer, BJ, R Ren, KL Clark & DS Baltimore (1993). A putative modular domain present in diverse signaling proteins. Cell 73(4): 629.<br> | |||
<br> | |||
Ren, R, BJ Mayer, P Cicchetti & D Baltimore (1993). Identification of a ten-amino acid proline-rich SH3 binding site. Science 259(5098): 1157.<br> | |||
<br> | |||
Lichtarge, O, HR Bourne & FE Cohen (1996 Mar 29). An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257(2): 342.<br> | |||
<br> | |||
Phizicky, E, PIH Bastiaens, H Zhu, M Snyder & S Fields (2003 Mar 13). Protein analysis on a proteomic scale. Nature 422(6928): 208.<br> | |||
<br> | |||
Bork, P & M Sudol (1994). The WW domain: a signalling site in dystrophin? Trends Biochem Sci 19(12): 531.<br> | |||
<br> | |||
Chen, HI & M Sudol (1995). The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A 92(17): 7819.<br> | |||
<br> | |||
Sudol, M & T Hunter (2000). NeW wrinkles for an old domain. Cell 103(7): 1001.<br> | |||
<br> | |||
Sudol, M (2005). WW Domain. in Modular Protein Domains. G Cesareni, M Gimona, M Sudol and M Yaffe. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim: 502.<br> | |||
<br> | |||
Gavin, AC, M Bosche, R Krause, P Grandi, M Marzioch, A Bauer, J Schultz, JM Rick, AM Michon, CM Cruciat, M Remor, C Hofert, M Schelder, M Brajenovic, H Ruffner, A Merino, K Klein, M Hudak, D Dickson, T Rudi, V Gnau, A Bauch, S Bastuck, B Huhse, C Leutwein, MA Heurtier, RR Copley, A Edelmann, E Querfurth, V Rybin, G Drewes, M Raida, T Bouwmeester, P Bork, B Seraphin, B Kuster, G Neubauer & G Superti-Furga (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868): 141.<br> | |||
<br> | |||
Ho, Y, A Gruhler, A Heilbut, GD Bader, L Moore, SL Adams, A Millar, P Taylor, K Bennett, K Boutilier, L Yang, C Wolting, I Donaldson, S Schandorff, J Shewnarane, M Vo, J Taggart, M Goudreault, B Muskat, C Alfarano, D Dewar, Z Lin, K Michalickova, AR Willems, H Sassi, PA Nielsen, KJ Rasmussen, JR Andersen, LE Johansen, LH Hansen, H Jespersen, A Podtelejnikov, E Nielsen, J Crawford, V Poulsen, BD Sorensen, J Matthiesen, RC Hendrickson, F Gleeson, T Pawson, MF Moran, D Durocher, M Mann, CW Hogue, D Figeys & M Tyers (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415(6868): 180.<br> | |||
<br> | |||
Bouwmeester, T, A Bauch, H Ruffner, PO Angrand, G Bergamini, K Croughton, C Cruciat, D Eberhard, J Gagneur, S Ghidelli, C Hopf, B Huhse, R Mangano, AM Michon, M Schirle, J Schlegl, M Schwab, MA Stein, A Bauer, G Casari, G Drewes, AC Gavin, DB Jackson, G Joberty, G Neubauer, J Rick, B Kuster & G Superti-Furga (2004). A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6(2): 97.<br> | |||
<br> | |||
Uetz, P, L Giot, G Cagney, TA Mansfield, RS Judson, JR Knight, D Lockshon, V Narayan, M Srinivasan, P Pochart, A Qureshi-Emili, Y Li, B Godwin, D Conover, T Kalbfleisch, G Vijayadamodar, M Yang, M Johnston, S Fields & JM Rothberg (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403(6770): 623. | |||
Ito, T, T Chiba, R Ozawa, M Yoshida, M Hattori & Y Sakaki (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A 98(8): 4569.<br> | |||
<br> | |||
Landgraf, C, S Panni, L Montecchi-Palazzi, L Castagnoli, J Schneider-Mergener, R Volkmer-Engert & G Cesareni (2004). Protein interaction networks by proteome peptide scanning. PLoS Biol 2(1): E14.<br> | |||
<br> | |||
Phizicky, E, PIH Bastiaens, H Zhu, M Snyder & S Fields (2003). Protein analysis on a proteomic scale. Nature 422(6928): 208.<br> | |||
<br> | |||
Tong, AH, B Drees, G Nardelli, GD Bader, B Brannetti, L Castagnoli, M Evangelista, S Ferracuti, B Nelson, S Paoluzi, M Quondam, A Zucconi, CW Hogue, S Fields, C Boone & G Cesareni (2002). A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295(5553): 321.<br> | |||
<br> | |||
Castagnoli, L, A Costantini, C Dall'Armi, S Gonfloni, L Montecchi-Palazzi, S Panni, S Paoluzi, E Santonico & G Cesareni (2004). Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett 567(1): 74.<br> | |||
<br> | |||
Songyang, Z & LC Cantley (2004). ZIP codes for delivering SH2 domains. Cell 116(2 Suppl): S41.<br> | |||
<br> | |||
Lee, CH, B Leung, MA Lemmon, J Zheng, D Cowburn, J Kuriyan & K Saksela (1995 Oct 16). A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J 14(20): 5006.<br> | |||
<br> | |||
Arold, S, R O'Brien, P Franken, MP Strub, F Hoh, C Dumas & JE Ladbury (1998 Oct 20). RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry 37(42): 14683.<br> | |||
<br> | |||
Marengere, LE, Z Songyang, GD Gish, MD Schaller, JT Parsons, MJ Stern, LC Cantley & T Pawson (1994). SH2 domain specificity and activity modified by a single residue. Nature 369(6480): 502.<br> | |||
<br> | |||
Hiipakka, M, K Poikonen & K Saksela (1999). SH3 domains with high affinity and engineered ligand specificities targeted to HIV-1 Nef. J Mol Biol 293(5): 1097.<br> | |||
<br> | |||
Zarrinpar, A, S-H Park & WA Lim (2003 Dec 11). Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426(6967): 676.<br> | |||
<br> | |||
Mayer, BJ (2001 Apr). SH3 domains: complexity in moderation. J Cell Sci 114(Pt 7): 1253.<br> | |||
<br> | |||
Cohen, IR (2001). Antigenic mimicry, clonal selection and autoimmunity. J Autoimmun 16(3): 337.<br> | |||
<br> | |||
Cohen, IR (2001). The creation of immune specificity. in Design Principles for Immune System & Other Autonomous Systems. LA Segel and IR Cohen. Oxford University Press, New York: 151–159.<br> | |||
<br> | |||
Parnes, O (2004). From interception to incorporation: degeneracy and promiscuous recognition as precursors of a paradigm shift in immunology. Mol Immunol 40(14-15): 985.<br> | |||
<br> | |||
}} | }} | ||
</div><br> | </div><br> | ||
== Who's visiting == | == Who's visiting == | ||
start on | <div style="padding: 10px; color: #ffffff; background-color: #000; width: 600px"> | ||
start on 18 Mar 2010 | |||
<html> | <html> | ||
<a href="http://clustrmaps.com/counter/maps.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" id="clustrMapsLink"><img src="http://clustrmaps.com/counter/index2.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" border= | <a href="http://www2.clustrmaps.com/counter/maps.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" id="clustrMapsLink"><img src="http://www2.clustrmaps.com/counter/index2.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onerror="this.onerror=null; this.src='http://clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://clustrmaps.com';" /> | ||
</a> | </a> | ||
</html> | |||
</div> | |||
==Science in the city: A small series about the everyday life of scientists in berlin== | ==Science in the city: A small series about the everyday life of scientists in berlin== | ||
'''FOLLOW ME''' [http://openwetware.org/wiki/User:Victor_Tapia#Science_in_the_city >>>>] | '''FOLLOW ME''' [http://openwetware.org/wiki/User:Victor_Tapia#Science_in_the_city >>>>] | ||
Revision as of 05:24, 28 February 2012
Contact Information
Molecular Recognition Laboratorium,
Institute für medizinische Immunologie
CHARITÉ - UNIVERSITÄTSMEDIZIN BERLIN

Hessische Str. 3-4 D-10115 Berlin, Germany phone +49-30-450 524092 fax +49-30-450 524942 mail annette.hayungs@charite.de web charite.de
last change on 01.04.2010
Group Leader
Group Members
- Annette Hayungs - Secretary
- Ines Kretzschmar - CTA
- Christiane Landgraf - CTA
- Judith Müller - Doktorandin
- Livia Otte - Postdoc
- Rolf-Dietrich Stigler - IT- u. Sicherheitsbeauftragter
- Víctor Tapia - Doktorand
- Lars Vouillème - Doktorand
Research interest
Technological Development of the Peptide Array Technologies
by Victor Tapia
FIGURE
PEPTIDE ARRAYS The combination of SPOT peptide synthesis (figure A, steps 1 to 4) with
appropriate immobilization techniques on glass supports (figure A, steps
5 and 6) is wide spread. The SPOT technology provides low-scale but
high-throughput synthesis, while immobilization of pre-synthesized
peptides offers the benefit of a "chemical" purification step and
flexible array design. Additionally, the glass support is compatible
with fluorescence detection (see figure B, adapted from the web) and
offers the possibility to miniaturize binding assays. Beyond economy,
the later point is essential for quantitative measurements at the
steady-state of binding activity, as has been described [Ekins 1998] and
can be proven by the mass-action law.
The basic point of this technology is the simultaneous display of a
systematic collection of peptides on a planar support, on which numerous
bimolecular interaction assays can be carried out under homogeneous
conditions.
MORE
PEPTIDE ARRAYS IN THE ADVANCEMENT OF PEPTIDE SYNTHESIS
- The development of solid-phase peptide synthesis (SPPS) by Bruce Merrifield [Gutte and Merrifield, 1969; Merrifield, 1965] and adaptions of this procedure [Fields and Noble, 1990] set the chemical ground for innovative technologies to follow.
- The development of the “Pin” method by H. Geysen [Geysen, et al., 1984] introduces the array format to peptide synthesis.
- Definitive establishment of peptide arrays came along with the development of the SPOT synthesis by Roland Frank [Frank, 1992; Frank, 2002] which simplified chemical synthesis of peptide arrays to the addressable deposition of reagents on a cellulose sheet.
Modern peptide synthesis approaches and
molecular biology make peptides accessible in a high degree of
structural diversity. The two greatest drawbacks of synthetic peptide
arrays are peptide length, with a quality threshold between 30 and 50
amino-acids, as well as the restriction to linear motives, since the
mimicry of nonlinear motives with linear peptide constructs is still
under development [Goede, et al., 2005].
PEPTIDE ARRAYS IN THE ADVANCEMENT OF BINDING ASSAY SYSTEMS
Since the 90s a major aspect of development to achieve the required
sensitivities to analyse biological samples has been the miniaturization
of analytical devices [Ekins, 1998]. It is important to note that
miniaturization is not only a matter of high-throughput and economy.
Miniaturization is an essential factor that should provide saturation of
binding sites under low analyte concentrations without significantly
altering its bulk (or ambient) concentration upon capturing [Ekins, et
al., 1990; Ekins, 1989; Joos, et al., 2002; Templin, et al., 2002].
- In this sense, the first application of a peptide microarray device in 1991, anticipating even the application of cDNA arrays, achieved already the impressive feature density of about 1024 peptides in 1.6 cm2 by means of in situ light-directed parallel synthesis [Fodor, et al., 1991].
Several methods available to generate peptide arrays on planar solid surfaces offer a range between...
- 16 peptides per cm2, in the case of SPOT macroarrays [Reimer, et al., 2002; Schutkowski, et al., 2004],
- to 2000-4000 peptides in 1.5 cm2, in the case of microarrays generated by digital photolithography [El Khoury, et al., 2007; Gao, et al., 2004; Pellois, et al., 2000; Pellois, et al., 2002].
SUPPORT MATERIALS
In order to
support synthesis, planar materials have to fulfil several requirements
including stability towards solvent and reagent deposition. The
functional groups on the surface must also be biochemically accessible
for chemical derivatisation. Furthermore, upon solid-phase binding
assay, generated peptides must be functionally displayed to allow
molecular recognition with a binding partner in the solution phase. In
particular non-specific interactions should be ruled out.
- Flexible porous supports such as cellulose [Eichler, et al., 1989; Frank and Döring, 1988], cotton [Eichler, et al., 1991; Schmidt and Eichler, 1993] or membranes [Daniels, et al., 1989; Wang and Laursen, 1992; Wenschuh, et al., 2000] are preferentially used for peptide array generation.
- Rigid, non-porous materials such as glass [Falsey, et al., 2001], gold films [Houseman and Mrksich, 2002; Jonsson, et al., 1991; Malmqvist, 1993], or silicon [Fodor, et al., 1991; Pellois, et al., 2002] have also been used for in situ synthesis, but are much more technically demanding.
- On the other side, rigid materials have a number of advantages over porous supports for functional display of molecules. Impermeability and smooth two-dimensionality of the material does not limit diffusion of the binding partner and leads to more accurate kinetics of recognition events.
- Finally the flatness and transparency of glass improve image acquisition and simplifies the use of fluorescence dyes for the read out process.
In some cases, assembled 3D structure on a non-porous surface could be
a fruitful approach. Several techniques for coherent surface modifications
are described over the past twenty years in the literature. For a comparative
overview on this field we refer to articles dedicated to the peptide and protein array
technologies [Angenendt and Glokler, 2004; Angenendt, et al., 2002;
Angenendt, et al., 2003; Seurynck-Servoss, et al., 2008;
Seurynck-Servoss, et al., 2007; Seurynck-Servoss, et al., 2007; Sobek,
et al., 2007; Sobek, et al., 2006; Wenschuh, et al., 2000].
REFERENCES
- Angenendt, P., and Glokler, J. (2004) Evaluation of antibodies and microarray coatings as a prerequisite for the generation of optimized antibody microarrays, Methods Mol Biol 264, 123-34.
- Angenendt, P., Glokler, J., Murphy, D., Lehrach, H., and Cahill, D. J. (2002) Toward optimized antibody microarrays: a comparison of current microarray support materials, Anal Biochem 309, 253-60.
- Angenendt, P., Glokler, J., Sobek, J., Lehrach, H., and Cahill, D. J. (2003) Next generation of protein microarray support materials: evaluation for protein and antibody microarray applications, J Chromatogr A 1009, 97-104.
- Daniels, S. B., Bernatowicz, M. S., Coull, J. M., and Köster, H. (1989) Membranes as solid supports for peptide synthesis., Tetrahedron Lett. 30.
- Eichler, J., Beyermann, M., and Bienert, M. (1989) Application of cellulose paper as support in simultaneous solid phase peptide synthesis., Colect. Czech. Chem. Commun. 54, 1746-52.
- Eichler, J., Bienert, M., Stierandova, A., and Lebl, M. (1991) Evaluation of cotton as a carrier for solid-phase peptide synthesis., Peptide Res. 4, 296-307.
- Ekins, R., Chu, F., and Biggart, E. (1990) Multispot, multianalyte, immunoassay, Ann Biol Clin (Paris) 48, 655-66.
- Ekins, R. P. (1989) Multi-analyte immunoassay, J Pharm Biomed Anal 7, 155-68.
- Ekins, R. P. (1998) Ligand assays: from electrophoresis to miniaturized microarrays, Clin Chem 44, 2015-30.
- El Khoury, G., Laurenceau, E., Dugas, V., Chevolot, Y., Merieux, Y., Duclos, M. C., Souteyrand, E., Rigal, D., Wallach, J., and Cloarec, J. P. (2007) Acid deprotection of covalently immobilized peptide probes on glass slides for peptide microarrays, Conf Proc IEEE Eng Med Biol Soc 2007, 2242-6.
- Falsey, J. R., Renil, M., Park, S., Li, S., and Lam, K. S. (2001) Peptide and small molecule microarray for high throughput cell adhesion and functional assays, Bioconjug Chem 12, 346-53.
- Fields, G. B., and Noble, R. L. (1990) Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids, Int J Pept Protein Res 35, 161-214.
- Fodor, S. P., Read, J. L., Pirrung, M. C., Stryer, L., Lu, A. T., and Solas, D. (1991) Light-directed, spatially addressable parallel chemical synthesis, Science 251, 767-73.
- Frank, R. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support, Tetrahedron, 9217-32.
- Frank, R. (2002) The SPOT-synthesis technique: Synthetic peptide arrays on membrane supports--principles and applications, J. Immunol. Methods 267, 13-26.
- Frank, R., and Döring, R. (1988) Simultaneous multiple peptide synthesis under continuous flow conditions on cellulose paper disks as segmental solid supports, Tetrahedron 44, 6031-40.
- Gao, X., Pellois, J. P., Na, Y., Kim, Y., Gulari, E., and Zhou, X. (2004) High density peptide microarrays. In situ synthesis and applications, Mol Divers 8, 177-87.
- Geysen, H. M., Meloen, R. H., and Barteling, S. J. (1984) Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid, Proc Natl Acad Sci U S A 81, 3998-4002.
- Goede, A., Jaeger, I. S., and Preissner, R. (2005) SUPERFICIAL--surface mapping of proteins via structure-based peptide library design, BMC Bioinformatics 6, 223.
- Gutte, B., and Merrifield, R. B. (1969) The total synthesis of an enzyme with ribonuclease A activity, J Am Chem Soc 91, 501-2.
- Houseman, B. T., and Mrksich, M. (2002) Towards quantitative assays with peptide chips: a surface engineering approach, Trends Biotechnol 20, 279-81.
- Jonsson, U., Fagerstam, L., Ivarsson, B., Johnsson, B., Karlsson, R., Lundh, K., Lofas, S., Persson, B., Roos, H., Ronnberg, I., and et al. (1991) Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology, Biotechniques 11, 620-7.
- Joos, T. O., Stoll, D., and Templin, M. F. (2002) Miniaturised multiplexed immunoassays, Curr Opin Chem Biol 6, 76-80.
- Malmqvist, M. (1993) Biospecific interaction analysis using biosensor technology, Nature 361, 186-7.
- Merrifield, R. B. (1965) Automated synthesis of peptides, Science 150, 178-85.
- Pellois, J. P., Wang, W., and Gao, X. (2000) Peptide synthesis based on t-Boc chemistry and solution photogenerated acids, J Comb Chem 2, 355-60.
- Pellois, J. P., Zhou, X., Srivannavit, O., Zhou, T., Gulari, E., and Gao, X. (2002) Individually addressable parallel peptide synthesis on microchips, Nat Biotechnol 20, 922-6.
- Reimer, U., Reineke, U., and Schneider-Mergener, J. (2002) Peptide arrays: from macro to micro, Curr Opin Biotechnol 13, 315-20.
- Schmidt, M., and Eichler, J. (1993) Multiple peptide synthesis using cellulose-based carriers: Synthesis of substance P - diastereoisomers and their histamine-releasing activity., Bioorg. Med. Chem. Lett. 3, 441-46.
- Schutkowski, M., Reimer, U., Panse, S., Dong, L., Lizcano, J. M., Alessi, D. R., and Schneider-Mergener, J. (2004) High-content peptide microarrays for deciphering kinase specificity and biology, Angew Chem Int Ed Engl 43, 2671-4.
- Seurynck-Servoss, S. L., Baird, C. L., Miller, K. D., Pefaur, N. B., Gonzalez, R. M., Apiyo, D. O., Engelmann, H. E., Srivastava, S., Kagan, J., Rodland, K. D., and Zangar, R. C. (2008) Immobilization strategies for single-chain antibody microarrays, Proteomics 8, 2199-210.
- Seurynck-Servoss, S. L., Baird, C. L., Rodland, K. D., and Zangar, R. C. (2007) Surface chemistries for antibody microarrays, Front Biosci 12, 3956-64.
- Seurynck-Servoss, S. L., White, A. M., Baird, C. L., Rodland, K. D., and Zangar, R. C. (2007) Evaluation of surface chemistries for antibody microarrays, Anal Biochem 371, 105-15.
- Sobek, J., Aquino, C., and Schlapbach, R. (2007) Quality considerations and selection of surface chemistry for glass-based DNA, peptide, antibody, carbohydrate, and small molecule microarrays, Methods Mol Biol 382, 17-31.
- Sobek, J., Bartscherer, K., Jacob, A., Hoheisel, J. D., and Angenendt, P. (2006) Microarray technology as a universal tool for high-throughput analysis of biological systems, Comb Chem High Throughput Screen 9, 365-80.
- Templin, M. F., Stoll, D., Schrenk, M., Traub, P. C., Vohringer, C. F., and Joos, T. O. (2002) Protein microarray technology, Drug Discov Today 7, 815-22.
- Wang, Z., and Laursen, R. A. (1992) Multiple peptide synthesis on polypropylene membranes for rapid screening of bioactive peptides., Pep. Res. 5, 275-80.
- Wenschuh, H., Volkmer-Engert, R., Schmidt, M., Schulz, M., Schneider-Mergener, J., and Reineke, U. (2000) Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides, Biopolymers 55, 188-206.
Messsystemanalyse von Peptidarray Techniken
by Victor Tapia
Mit dem Ziel die Affinitätsvorhersagbarkeit von Peptidarray-orientierten Bindunsstudien zu analyzieren, wurde die CB4 1 Epitope Bibliothek definiert, welche auf ein Repertoire von 26 verschiedene Liganden für das CB4-1 Antikörper besteht. Es handelt sich hierbei haupsächlich um Sequenz- und Mimika¬varianten des natürlichen Epitopes von CB4-1 (? p24 von HIV 1). Die Eignung dieser Proben für das oben erklärtes Ziel beruhrt auf das vorhandene Affinitätsprofil, das jedes Peptid mit einzeln gemessenen KD Werte assoziiert.
Die einzelne KD Messungen wurden teilweise aus der Literatur aussortiert [Volkmer-Engert 1995; Kramer 1997; Kramer 1998; Hoffmuller 2000; Reineke 2002] und teilweise mit eigenen SPR Messungen belegt [Weiser 2005].
Das Peptid Repertoire der Bibliothek umfasst ein Affinitätsbereich mit KD-Werten zwischen 1 mM und 1 nM. Diese Bibliothek wird sowohl zur Analytbestimmung wie auch zur Affinitätsvorhersage eingesetzt, um die Bediengungen eines Assays zu determinieren, unter welcher eine wertgenaue Vorhersage möglich ist.
Hierbei handelt es sich um ein Bindungsstudie in Mikroarray-Format [Reimer 2002; Tapia 2009]. Die Wert¬genauig¬keit der Ansatz wird anhand der Übereinstimmung mit Referenz Affinitätswerten (Sollwerte) aus den einzelne SPR-bezogene Bindunsstudien. Der Einsatz
dieser Referenzmesswerte als „goldener Standard“ ist begründet durch drei wessentliche unterschiede zu den Mikro- und Makroarray Formate der HTS Bindungsstudien: (1) die Peptid Proben sind ausführlich präpariert worden (HPLC, MS), (2) das Analyt wurde durch die Messzelle des Sensorchips durchflossen und (3) die Auswertung geschieht anhand von Sensogrammen, die eine echtzeit Beobachtung der Gleichgewichtzustandes ermöglicht (siehe Abbildung 1 für ein Überblick).
MORE TEXT 1
Die 26 Epitopvarianten der CB4-1 Epitopebibliothek wurden als Amino-oxyacetylierte Peptid-Proben via Spot Technik synthetiziert. Nach der Abspaltung der Schutzgruppen der Seitenkette wurden die Proben auf eine Mikrotitterplatte transferiert. Anschlißend, wurden die Rohprodukten der Proben auf Aldehyd-funktionalizierte Glass-Trägern mit einem piezoelektrischen Printer immobiliziert. Dadurch wurden die Peptide von N- (gebunden) zu C-Term (frei) präzentiert. Die Arrayanordnung ist in Abbildung 2 dargestellt. Für die vier Wiederholungen der dort gezeigten Bindunsstudie, stammten die funktionalizierte Plattformen aus einem Produktions-„batch“ des Herstellers (Genetix Ltd., UK) und ebenso wurden in einem „batch“ darauf die Peptidproben immobiliziert und mit dem CB4 1 Analyt inkubiert. Ein Minimum an technische Quellen für Variation in den Messungen wurde in dieser Weise angestrebt.
Abbildung 2. A.- Die vier Spalten zeigen die Negativ-Verarbeitung der Bildaufzeichnungen von vier Wiederholungen eines Mikroarray Bindungsstudie der CB4-1 Antikörper auf 20 Replikas eines 4?8 Arrays (Panel C). Die 20 Arrays jedes Plattforms sind vertikal in fünf Felder von 4 Arrayreplikas angeordnet. B.- Vergrößerung des viereckigen Rahmen im Panel A, d.h. ein Replikafeld. Die aufgezeichneten Signalintensitäten sind, soweit unspezifische Wechselwirkungen des Detektionsantikörpers ausgeschlossen werden konnten, von den Mengen an Analyt abhängig, die auf jeden Peptidspot eingefangen wurden. C.- Die Positionen des 4?8 Arrays wurden mit den Peptidsequenzen der Subbibliothek besetzt. Die angegebene Zahlen auf jeden Spot entsprechen die interne ID-Erkennungszeichen aus Tabelle 1. Die Sequenzspezifität des Immunoanalyts (?-p24 HIV-1, CB4-1) kann aus der Zusammenschluß beider Informationen, d.h. positionsabhängige Signalintensitäten und adressierbare Sequenzliste, abgeleitet werden. Die Abbildung zeigt insgesamt 80 Wiederholungen von Proben 1, 3-17, und 19-26 sowie 320 Wiederholungen von Proben 2 (schwach Binder) und 18 (stark Binder).
Die Qualität der Fluoreszenzmessungen wurde in zwei Hinsichten
untersucht: Präzision und Wertgenauigkeit. Die Präzision der Technik
wurde mittels eine beschreibende Untersuchung der Variation in der
Messung charakterisiert. Dabei wurde die Streuung von Signalintensitäten
aus Replikaspots, die innerhalb eines Plattforms verteilt waren, durch
das Variations¬koeffizient (engl. „coefficient of variation“, COV=
Standard Abweichung/Mittelwert) erfasst. Im Durchschnitt, ergibt das COV
der vier Plattformen ein Wert von 0,31 ± 0,177 (Abbildung 3, Panel A).
Dieser Wert entspricht die Streuung der Signalintensität
(Wiederholpräzision, engl. repeatability), die bei äquivalenten
Messungen innerhalb eines Plattforms erwartet wird. Wiederum, der
paarweise Vergleich der kompleten Datensätze jedes Plattforms mit
einander durch lineare Korrelationsanalyse ist in der Lage die
Nachvollziehbarkeit (Vergleichspräzision, engl. reproducibility) der
gemessene Signalintensitäten zu beschreiben. Im Durchnitt, das
Determinationskoeffizient der 6 nicht-redundanten paarweisen Vergleiche
ergibt 0,98 ± 0,013 (Abbildung 3, Panel B). Dieser Wert beschreibt eine
technische Fähigkeit, indem von 100 Vergleiche 98 nicht unterscheidbare
Messungen zu erwarten sind. Eine Erweiterung dieser Untersuchung auf 3
weitere unterschiedliche Plattform Produkte und 2 Liefermengen (engl.
batches) wurde in [Tapia 2007] veroffentlicht.
Abbildung 3. Panel A: Wiederholpräzision des Mikroarray-Messsystems. Die Fähigkeit, ein kohärentes Mittelwert von Replikas innerhalb einer Messung zu wiederspiegeln, ist für vier Plattforms (MS1-4), die wie im Abbildung 2 mit Peptid Proben belegt wurden, beschrieben. Der globale Durchschnitt der COV Werte (gestrichelte Linie) ist 0,31 ± 0,177. Panel B: Vergleichspräzision des Mikroarray-Mess¬systems. Die Fähigkeit, kohärente Messergebnisse mit unabhängigen Messungen zu erzielen, ist durch die Bestimmtheitsmaß (R2) von paarweisen Vergleichen der Fluoreszenz (engl. mean SI) beschrieben. Im Durchschnitt R2=0,98 ± 0,013.
Die Wertgenauigkeit (oder Richtigkeit, engl. accuracy) der Signalmessungen wird mittels einer analytischen Untersuchung zweierlei Effekten auf die von Arrayproben eingefangene Analytmenge beschrieben: erstens, der Effekt von variierenden Affinitäts¬werte (Analyt-bestimmung) und, zweitens, der Effekt von variierenden Analyt¬konzentrationen (Affinitäts-bestimmung). Hierbei unterscheidet man zwischen Soll- und Istwerte (engl. „nominal“ und „actual values“, entsprechend). Bei der Analytbestimmung ist die experimentell justierbare Variable durch unabhängig bestimmte Affinitäten der Peptidproben repräsentiert. Für zwecken der Analyse wird der Einfluß dieser Variable auf die eingefangene Analytmenge (und dadurch auf die gemessene Signalintensitäten) bei eine Serie von unterschiedlich eingesetzten Analytkonzentrationen untersucht. Die Sollwerte der Analytbestimmung sind somit durch die fünf verwendete molare Konzentrationen (cAN={3,34 µM; 667 nM; 66,7 nM; 6,67 nM und 667 pM}) vertretten.
Tabelle 3. Vergleich zwischen beider Ansätze der Vorhersage.
Ansatz experimentell justierbare Variable Istwert Sollwert Model
Analytbestimmung 26 pKD Werte cA cAN ?=?(pKD)
Affinitätsbetimmung 5 cA Werte EC50 pKDN ?=?(cA)
Dagegen, ist bei der Affinitätsbestimmung die experimentell justierbare Variable durch eine Reihe von bestimmten Analytkonzentrationen repesentiert. Für zwecken der Analyse wird der Einfluß dieser Variable bei eine Serie von unabhängig gemessenen Affinitäten untersucht. Die Sollwerte der Affinitätsbestimmung sind somit durch die 26 verwendete Affinitäten (pKDN aus Tabelle 1) vertretten.
Die Istwerte (engl. actual values) werden durch die geschätzte Werte, die mit der jeweiligen Lösungsansätzte der selber Regressionsmodel erzielt wurden, representiert. Die Analytbestimmung ergibt sich aus der Formulierung der Regressionsmodel als ?=?(pKD) mit dem Systemkonstante cA. Für jeder Inkubation werden die cA als Istwerte geerntet. Durch eine Reformulierung des Regressionsmodels als ?=?(cA) wird bei der Affinitätsbestimmung das Systemkonstanten KD jeder Peptidprobe als Istwert geerntet.
Abbildung 4. Wertgenauigkeit der Analytbestimmung. Hier handelt es sich um ein Gleichgewichtsanalyse der Effekt von variierten Affinitätswerten (einzelne SPR pKD Werte) auf die Analytmenge, die von den Arrayproben eingefangen werden. Panel A und B zeigen die Streuung der SI Messdaten und die Regressionskurven, die auf unabhängige Inkubationen der Arrayproben mit einer 3,34 µM bzw. einer ca. 0,67 nM Analytkonzentration (cAN Sollwert, engl. nominal value), entsprechend, zurückzuführen sind. Panel C fasst die Regressionskurven für alle angewandte Analyt¬konzentrationen (cAN= {3,3 µM; 670 nM; 67 nM; 6,7 nM und 0,67 nM}) zusammen. Die dazu gehörige Parameterwerte (cA Istwerte, engl. actual cA) sind im Panel D mit cAN. Zur Diskussion der Kurvenverläufen in Panel D ist außerdem dem geschätzten Wert der Peptidkonzentration (cP Istwert, engl. cP actual value) vergleichend dargestellt.
Abbildung 4 fasst die Ergebnisse der Analytbestimmung zusammen. Panel A und B beweisen einen mit der Messdaten koherente Verlauf der Regressionskurven für zwei der angewandeten Analytkonzentrationen, 3,34 µM und 0,67 nM entsprechend. Ebenso in Panel C ersichtlich ist ein mit den angewandten Analytkonzentrationen koherenten Verrückung der Regressionskurven. Auf dem ersten Blick scheint die niedrigste Konzentration (0,67 nM) dieser Beobachtung zu wiedersprechen. Dafür werden in Panel D die Werte der Analytbestimmung mit dem geschätzten Wert der Peptidkonzentration verglichen. Offensichtlich, ist im Messsystem eine 0,67 nM Konzentration des Analyts niedriger als die Peptidkonzentration. Dieser Zustand führt zu einer im Gleichgewicht unvollständigen Besetzung der Bindungstellen. Einer Erhöhung der Analytkonzentration muss zuerst die vollständige Besetzung der Bindungstellen bewirken bevor die Regressionskurven höhere Analytkonzentrationen entsprechend nach links im Panel C verrücken können. Dieser Möglichkeit soll während des fitting-Verfahrens unbedingt beachtet werden.
Die Wertgenauigkeit der Analytbestimmung ist durch eine paarweise lineare Korrelation der Ist- und Sollwerte der Analytkonzentrationen mit einem Bestimmtheitsmaß R2=0.98 charakteriziert. Allerdings, eine systematische Messabweichung zu niedrigen Istwerte bei Sollwerte > 67 nM und zu höheren Istwerte bei Sollwerte < 67 nM ist ersichtlich. Es ist aus dieser Analyse nicht zu entnehmen, ob eine Erwieterung der angewendten Anaylt-konzentrationen mit extremeren Werten zu einer Bestätigung oder Ausgleich der systematischen Abweichung führen kann.
Von unmittelbare Interesse für die gegenwärtige Arbeit is die Bestimmung von Affinitätswerte. Solches Ziel gelingt durch die Untersuchung der Abhängigkeit der eingefangenen Analytmenge von der Analytkonzentration (justierbare Variable). Hierbei handelt es sich um die statistische Schätzung (oder, etwas optimitischer, die Vorhersage) der Systemkonstanten (insbesondere Istwerte, negativen dekadischen Logarithmus der Dissoziations¬konstante KD in mol*L-1). Die Genauigkeit der Vorhersage lässt sich durch lineare Korrelation mit dem Affinitätssollwerten (negativen dekadischen Logarithmus aus unabhängigen einzelne KDN Messungen in mol*L-1) bestimmen.
Abbildung 5 fasst die Ergebnisse der Affinitätsbestimmung in Analogie zu Abbildung 4 zusammen. Panel A und B zeigen die kohärenz der Kurvenverläufe in Bezug auf die Verteilung der Messdaten mit Bestimheitsmaßen von R2=0,98 (schwacher Binder, pKDN= 3,52) und 0,95 (starker Binder, pKDN= 8,22), entsprechend. Um die Abhängigkeit der SI von variirenden Analytkonzentrationen darzustellen, wurde die Anordnung der Datensätze im Vergleich zu Abbildung 4 transponiert. <br>
Dadurch findet in Panel A bis C ein Kurvenverlauf von einem SI Minimum zu einem Maximum in Umgekehrte Richtung statt. Panel C zeigt die durch die sinkende Affinitätsistwerte definierte links-Verrückung der Kurvenverläufen. Die Regressionskurven erreichen asymptotisch ein global definiertes SI Maximum.
Im Panel D von Abbildung 5 ist eine kaum lineare Vergleich der Messdaten ersichtlich (R2?0,70). Zwei Aspekte verdienen besondere Kritik und werden die Versuchsplannung der PQBP-1 Bindungsstudien entscheidend Prägen. Erstens, Sollwerte zwischen 3,5 und 6,2 werden von Istwerte begegnet, die zwischen 4,2 und 5.8 einengent bestimmt wurden. Zweitens der schmalen dynamischen Bereich bei Sollwerten von 6,2 bis etwa 7,3 wird von kaum korrelierenden Istwerten begegnet (R2<0,50).
Abbildung 5. Wertgenauigkeit der Affinitätsvorhersage. Hier handelt es sich um ein Gleichgewichtsanalyse der Effekt von variierten Analytkonzentrationen auf die Analytmenge die von den Arrayproben eingefangen werden. Panel A und B zeigen die Streuung der SI Messdaten und die Regressionskurve für die Inkubation eines belegten Plattforms mit einer etwa 3 µM und einer etwa 0,7 nM Analytkonzentration (cA Sollwert, engl. nominal value), entsprechend. Panel C fasst die Regressionskurven für alle angewandte Analytkonzentrationen zusammen. Die dazu gehörige Parameterwerte (cA Istwert, engl. actual value) sind im Panel D mit dem erwarteten cA Sollwert und dem geschätzten Wert der Peptidkonzentration (cP Istwert, engl. cP actual value) vergleichend dargestellt.
Structural Modularity in Protein-Protein Recognition
by Victor Tapia
FIGURE
Protein Interaction Domains
The organisation of living systems is a complex network of molecular interactions. Proteins are a central component of the network as they may bind to other proteins as well as to phospholipids, nucleic acids and small molecules to interconnect the diverse physiological functions of the cell. In the background of these observations, the existence of a molecular recognition code for cellular organisation is very suggestive.
MORE TEXT
Structural analysis of functional protein complexes suggests at least two classes of protein-protein interactions. In the first class, the so called surface/surface interactions, complementary rigid surfaces link the interacting partners. Under these circumstances, the residues involved in each interacting surface only come together upon protein folding. The second class consists on asymmetric or surface/string interactions, where a protein domain (folding module of moderate size that may resemble a pocket on the protein’s surface) docks a short lineal peptide motive (a peptide ligand) on the partner protein. While surface/surface interactions cannot be inferred because the primary structure is not linearly involved, the binding determinants of a protein interaction domain (PID) may be mapped to short peptides matching the sequence of the ligand peptide. The importance of these recognition domains in the formation of protein complexes by binding to short lineal peptides was firstly demonstrated in the late 1980s and early 1990s [Sadowski 1986 Dec; Mayer 1993; Ren 1993].
In a pioneering work on the kinase function and transforming activity of the Fujinami Sarcoma Virus, Sadowski et al.[Sadowski 1986 Dec] discovered “a unique domain… (which) is absent from kinases that span the plasma membrane” and concluded that “the presence of this noncatalytic domain in all known cytoplasmic tyrosine kinases of higher and lower eukaryotes argues for an important biological function... the noncatalytic domain may direct specific interactions of the enzymatic region with cellular components that regulate or mediate tyrosine kinase function”. These regions were called Src homology 2 (SH2) and 3 (SH3), the name SH1 being reserved to the catalytic region. Since then, the gained knowledge on SH2/3 domain function has been a paradigm in our understanding of PID biochemistry, e.g. PIDs as non-catalytic binding sites and with modular structure.
In this era of extensive genome sequencing, many PIDs have been discovered. The interaction partners — and, therefore, the functions of many proteins ― may be determined by identifying the critical binding sites for family members through evolutionary tracing [Lichtarge 1996 Mar 29] or mapping protein-protein interactions using functional protein arrays [Phizicky 2003 Mar 13]. Many of the PIDs in proteins can be grouped into families that show clear evidence of their evolution from a common ancestor, and genome sequences from Saccharomyces cerevisiae to Homo sapiens reveal large numbers of proteins that contain one or more common domain.
Modularity of WW PIDs
The WW domain is a well-characterized protein module that mediates specific protein-protein interactions with ligands that contain short proline-rich motifs [Bork 1994; Chen 1995; Sudol 2000]. The domain is composed of 38 amino acids, it has two signature tryptophan (W) residues, and it is one of the smallest among modular protein domains. The signature tryptophan positions are spaced 20–22 amino acids apart and have important structural role. The structure is a three beta-strand meander that forms a shallow binding pocket for proline-rich ligands (reviewed in[Sudol 2005]). In general, the folding of WW domains does not require cognate ligands or co-factors.
MORE TEXT
There are two aspects which allow a protein motiv to be recognized as module. Firstly, functional modularity: a domains ability to function independent of context, allowing transfer of function between diverse molecular systems. In the case of a WW domain, it functionally resembles the Src homology domain 3 in its recognition of proline-rich or proline-containing ligands. Based on this ligand predilection, two major and two minor groups are classified:
Group I core consensus PPxY Group II PPLP motif Group III poly-P motifs flanked by R or K Group IV short seq. with pS or pT followed by P
The involved affinity of interactions in terms of KD for polyproline rich ligands (Groups I to III) range between µM to nM values. For pSP- or pTP-containing ligands, binding strengths are measured in the low µM range.
Secondly, structural modularity: the ability of a protein motiv to be physically isolated from its protein context and yet fold into the characteristic conformation (structural autonomy). Although several WW domain structures are annotated in the Protein Database (PDB), some can only be shown to fold stabily upon ligand binding. It is a matter of controversy, if ligand binding induces the fold or if it selects a stable bounded conformation from many dynamically interchanging free conformations. This is analogous to the question whether ligand binding or folding firstly occurs.
From Promiscuous Recognition Events to Mutually Exclusive Cellular Responses
The elucidation of functional pathways of signal transduction, biochemical function or gene regulation, is firstly addressed in proteomics by deriving interaction networks depicting ideally all interactions in the cell. Several attempts have been done in this direction on different model organisms and with varied methods, including co-purification by affinity chromatography [Gavin 2002; Ho 2002; Bouwmeester 2004], yeast two-hybrid [Uetz 2000; Ito 2001], phage display, peptide array technologies [Landgraf 2004], etc. A comparison of datasets derived by individual methods demonstrates that different methods have different potential.
MORE TEXT
For example, affinity chromatographic approaches are biased to tight interactions such as those involving extensive complementary surfaces, while interactions in which one of the two partners contains at least one PID are more frequent in the two-hybrid database. The higher sensitivity of the so called synthetic approaches (yeast two-hybrid, phage display and peptide array technologies) make them better suited for detecting PID-mediated interactions since their peptide affinity in terms of KD falls in the 10 – 100 µM range (high Kd --> low affinity). However, this advantage is counterbalanced by a low specificity, especially of the yeast two-hybrid approach [Phizicky 2003].
In order to correct this deficiency a double check-up of the information fed into the interaction databases is recommended. This can be achieved by deriving two interaction networks through orthogonal (fundamentally different) synthetic methods and then considering only the intersection between the two datasets [Tong 2002; Castagnoli 2004; Landgraf 2004]. False positive reports are thus reduced if the causes for measurement error are different in each method.
The strength of this combined approach to deliver physiologically relevant interactions has been proven for a phage display/yeast two-hybrid intersected dataset [Tong 2002]. A notable conclusion of this approach is that the intersected dataset of proteins that are able to interact with a given PID is larger than expected when cellular events are viewed as a precise wiring of the proteins in the cell. Although a set of these biochemically potential binders may have no physiological relevance due to expression at different times or tissues, in vitro disrupted structures, etc., the paradox of promiscuous recognition and mutually exclusive responses seems to be inherent to PID mediated interactions: the work of Landgraf et al. [Landgraf 2004] supports the observation that a large fraction of natural peptides with the biochemical potential to bind to any given SH3 domain is actually used in vivo to mediate the formation of a complex.
An additional difficulty to derive functional interaction pathways is that the difference in affinity between ‘specific’ and ‘non-specific’ interactions has been shown to be less than two orders of magnitude in the case of SH2 and its peptide ligands [Songyang 2004]. Even when granted that the recognition specificity of intact proteins by SH3 domains is greater than for SH3-peptide recognition, affinity is not raised above one order of magnitude [Lee 1995 Oct 16; Arold 1998 Oct 20]. Moreover, the ability of a point-mutant Src SH2 domain to effectively substitute for the SH2 domain of the Sem-5 protein in activation of the Ras pathway in vivo emphasises that the specificity of Sh2-mediated interactions is not great [Marengere 1994]. Consider the later statements under the light of the fact that the affinity of the protein OppA for its ligands is in the range of two orders of magnitude (Oppa is involved in the mopping of peptides in the bacterial periplasm exhibiting no sequence specificity). Tu put it all into a nutshell: the described facts lead to a view of large and promiscuous SH-mediated interaction networks.
Since it is possible to generate mutant SH3 domains that have up to 40-fold higher affinity than their wild-types [Hiipakka 1999] , the potential of these domains as research tools and as source of lead compounds for pharmaceutical development can not be overseen. Furthermore, a question cannot be overheard in our minds: which is the functional advantage of maintaining relative low affinity and selectivity for PID-mediated interactions, instead of optimizing the potential affinity of PIDs? And further: how can PID-dependent interaction pathways achieve precise cellular responses?
A comfortable view is that sufficient effective selectivity can be brought by compartmentalization, additive effects of multiple separate interactions, cooperative assembly of multiprotein complexes, etc. and that these effects can sustain linear functional pathways. An interesting insight into this mater has been advanced by Zarrinpar, A. et al. In their work [Zarrinpar 2003 Dec 11] the authors find out that while metazoan SH3 domains may rescue the functionality of mutated Sho1-SH3 of the yeast, in the set of yeast-own SH3 domains this promiscuity is forbidden. They thus conclude and confirm that, on the background of diverging SH3 domains, negative selection has drifted the ligand sequences to non-overlapping areas of the particular SH3 binding regions on the sequence space.
Alternatively, a divergence process of the shape of the binding region may be ‘guided’ by positive selection to avoid overlapping and, thus, promiscuitive interactions. Nevertheless, the picture of linear functional pathways is being revolutionized by a more probabilistic view of a dynamical equilibrium between multiple interactions, in which “the central organizing principle is a vast and ever-shifting web of interactions, from which output is gauged by global changes in complex binding equilibria”[Mayer 2001 Apr]. Cellular systems are explained here as transformational systems in which a redundant or degenerate input is transformed into a meaningful pattern. Irun Cohen’s ‘emergent specificity’, as postulated to synthesize the clonal selection theory and our knowledge on the promiscuity of immunoreceptors[Cohen 2001; Cohen 2001; Parnes 2004], aims at stitching this fissure with nonlinearity. Emergent properties are those behaviours manifested by a system when it operates as a whole, with no straightforward reducibility to the particulate components taking part in the process.
REFERENCES
Sadowski, I, JC Stone & T Pawson (1986 Dec). A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol 6(12): 4396.
Mayer, BJ, R Ren, KL Clark & DS Baltimore (1993). A putative modular domain present in diverse signaling proteins. Cell 73(4): 629.
Ren, R, BJ Mayer, P Cicchetti & D Baltimore (1993). Identification of a ten-amino acid proline-rich SH3 binding site. Science 259(5098): 1157.
Lichtarge, O, HR Bourne & FE Cohen (1996 Mar 29). An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257(2): 342.
Phizicky, E, PIH Bastiaens, H Zhu, M Snyder & S Fields (2003 Mar 13). Protein analysis on a proteomic scale. Nature 422(6928): 208.
Bork, P & M Sudol (1994). The WW domain: a signalling site in dystrophin? Trends Biochem Sci 19(12): 531.
Chen, HI & M Sudol (1995). The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A 92(17): 7819.
Sudol, M & T Hunter (2000). NeW wrinkles for an old domain. Cell 103(7): 1001.
Sudol, M (2005). WW Domain. in Modular Protein Domains. G Cesareni, M Gimona, M Sudol and M Yaffe. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim: 502.
Gavin, AC, M Bosche, R Krause, P Grandi, M Marzioch, A Bauer, J Schultz, JM Rick, AM Michon, CM Cruciat, M Remor, C Hofert, M Schelder, M Brajenovic, H Ruffner, A Merino, K Klein, M Hudak, D Dickson, T Rudi, V Gnau, A Bauch, S Bastuck, B Huhse, C Leutwein, MA Heurtier, RR Copley, A Edelmann, E Querfurth, V Rybin, G Drewes, M Raida, T Bouwmeester, P Bork, B Seraphin, B Kuster, G Neubauer & G Superti-Furga (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868): 141.
Ho, Y, A Gruhler, A Heilbut, GD Bader, L Moore, SL Adams, A Millar, P Taylor, K Bennett, K Boutilier, L Yang, C Wolting, I Donaldson, S Schandorff, J Shewnarane, M Vo, J Taggart, M Goudreault, B Muskat, C Alfarano, D Dewar, Z Lin, K Michalickova, AR Willems, H Sassi, PA Nielsen, KJ Rasmussen, JR Andersen, LE Johansen, LH Hansen, H Jespersen, A Podtelejnikov, E Nielsen, J Crawford, V Poulsen, BD Sorensen, J Matthiesen, RC Hendrickson, F Gleeson, T Pawson, MF Moran, D Durocher, M Mann, CW Hogue, D Figeys & M Tyers (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415(6868): 180.
Bouwmeester, T, A Bauch, H Ruffner, PO Angrand, G Bergamini, K Croughton, C Cruciat, D Eberhard, J Gagneur, S Ghidelli, C Hopf, B Huhse, R Mangano, AM Michon, M Schirle, J Schlegl, M Schwab, MA Stein, A Bauer, G Casari, G Drewes, AC Gavin, DB Jackson, G Joberty, G Neubauer, J Rick, B Kuster & G Superti-Furga (2004). A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6(2): 97.
Uetz, P, L Giot, G Cagney, TA Mansfield, RS Judson, JR Knight, D Lockshon, V Narayan, M Srinivasan, P Pochart, A Qureshi-Emili, Y Li, B Godwin, D Conover, T Kalbfleisch, G Vijayadamodar, M Yang, M Johnston, S Fields & JM Rothberg (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403(6770): 623.
Ito, T, T Chiba, R Ozawa, M Yoshida, M Hattori & Y Sakaki (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A 98(8): 4569.
Landgraf, C, S Panni, L Montecchi-Palazzi, L Castagnoli, J Schneider-Mergener, R Volkmer-Engert & G Cesareni (2004). Protein interaction networks by proteome peptide scanning. PLoS Biol 2(1): E14.
Phizicky, E, PIH Bastiaens, H Zhu, M Snyder & S Fields (2003). Protein analysis on a proteomic scale. Nature 422(6928): 208.
Tong, AH, B Drees, G Nardelli, GD Bader, B Brannetti, L Castagnoli, M Evangelista, S Ferracuti, B Nelson, S Paoluzi, M Quondam, A Zucconi, CW Hogue, S Fields, C Boone & G Cesareni (2002). A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295(5553): 321.
Castagnoli, L, A Costantini, C Dall'Armi, S Gonfloni, L Montecchi-Palazzi, S Panni, S Paoluzi, E Santonico & G Cesareni (2004). Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett 567(1): 74.
Songyang, Z & LC Cantley (2004). ZIP codes for delivering SH2 domains. Cell 116(2 Suppl): S41.
Lee, CH, B Leung, MA Lemmon, J Zheng, D Cowburn, J Kuriyan & K Saksela (1995 Oct 16). A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J 14(20): 5006.
Arold, S, R O'Brien, P Franken, MP Strub, F Hoh, C Dumas & JE Ladbury (1998 Oct 20). RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry 37(42): 14683.
Marengere, LE, Z Songyang, GD Gish, MD Schaller, JT Parsons, MJ Stern, LC Cantley & T Pawson (1994). SH2 domain specificity and activity modified by a single residue. Nature 369(6480): 502.
Hiipakka, M, K Poikonen & K Saksela (1999). SH3 domains with high affinity and engineered ligand specificities targeted to HIV-1 Nef. J Mol Biol 293(5): 1097.
Zarrinpar, A, S-H Park & WA Lim (2003 Dec 11). Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426(6967): 676.
Mayer, BJ (2001 Apr). SH3 domains: complexity in moderation. J Cell Sci 114(Pt 7): 1253.
Cohen, IR (2001). Antigenic mimicry, clonal selection and autoimmunity. J Autoimmun 16(3): 337.
Cohen, IR (2001). The creation of immune specificity. in Design Principles for Immune System & Other Autonomous Systems. LA Segel and IR Cohen. Oxford University Press, New York: 151–159.
Parnes, O (2004). From interception to incorporation: degeneracy and promiscuous recognition as precursors of a paradigm shift in immunology. Mol Immunol 40(14-15): 985.
Who's visiting
start on 18 Mar 2010
<html> <a href="http://www2.clustrmaps.com/counter/maps.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" id="clustrMapsLink"><img src="http://www2.clustrmaps.com/counter/index2.php?url=http://openwetware.org/wiki/Molecular_Recognition_Laboratorium" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onerror="this.onerror=null; this.src='http://clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://clustrmaps.com';" /> </a> </html>
Science in the city: A small series about the everyday life of scientists in berlin
FOLLOW ME >>>>