Maloof Lab:Jose M. Jimenez-Gomez: Difference between revisions
No edit summary |
No edit summary |
||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
<h2>Jose M Jimenez-Gomez, PhD.</h2> | <h2>Jose M Jimenez-Gomez, PhD.</h2> | ||
[mailto:jmjimenez@ucdavis.edu Contact] | [mailto:jmjimenez@ucdavis.edu Contact] | ||
<p> | |||
<br> | |||
<font color='red'>The information contained in this website may be outdated.</font><br> | |||
Please use my this website instead: [http://jimenez-gomez_lab.openwetware.org Jimenez Gomez Lab]. | |||
<br> | |||
</p> | |||
|[[Image:Pepe_b&w.jpg|right|135px]] | |[[Image:Pepe_b&w.jpg|right|135px]] | ||
|} | |} | ||
<br> | <br> | ||
I am a Postdoctoral fellow in [[Maloof_Lab |Julin Maloof's lab]] in the [http://www-plb.ucdavis.edu/ Section of Plant Biology] at the [http://www.ucdavis.edu University of California Davis].<br> | Starting in October 2010, I am a Junior Group Leader in the Department of Plant Breeding and Genetics at the [http://www.mpiz-koeln.mpg.de Max Planck Institute for Plant Breeding] in Cologne. | ||
I worked as a Postdoctoral fellow in [[Maloof_Lab |Julin Maloof's lab]] in the [http://www-plb.ucdavis.edu/ Section of Plant Biology] at the [http://www.ucdavis.edu University of California Davis].<br> | |||
<br> | <br> | ||
In 2005, I completed my PhD. in JM Martinez-Zapater's lab at the [http://www.cnb.uam.es CNB] (National Center for Biotechnology) in Madrid, Spain, where I performed a quantitative genetic analysis of flowering time in tomato <cite>Jimenez-Gomez07</cite>. | In 2005, I completed my PhD. in JM Martinez-Zapater's lab at the [http://www.cnb.uam.es CNB] (National Center for Biotechnology) in Madrid, Spain, where I performed a quantitative genetic analysis of flowering time in tomato <cite>Jimenez-Gomez07</cite>.<br> | ||
<br> | |||
My main interests are based on the application of modern genetic and bioinformatic techniques to the study of plant natural variation, evolution and domestication. To do this I survey different plant species and populations presenting variation in interesting characteristics, and analyze the responsible molecular mechanism. Here is an small description of some of my recent work in the Maloof lab: | |||
<br> | <br> | ||
<br> | <br> | ||
<h3><font style="color:#F8B603;">QTL analysis of the shade avoidance response in Arabidopsis</font></h3> | <h3><font style="color:#F8B603;">QTL and Network analysis of the shade avoidance response in Arabidopsis</font></h3> | ||
---- | ---- | ||
<br> | <br> | ||
It is well known that plants from different light environments exhibit different degrees of responsiveness to similar light stimulus. For example, plants accommodated to sunny environments detect foliar shade from neighboring vegetation and respond increasing their petioles/stems and reducing the time to reproduction, a phenomenon called the "shade avoidance response". On the other hand, plants adapted to live under dense canopies are less sensitive to the shade and have a reduced shade avoidance response. | |||
To identify the molecular mechanisms underlying this differences we are performing QTL analysis using a previously developed, well characterized Recombinant Inbred Line set descent from two different natural populations of <i>Arabidopsis thaliana</i>: Bayreuth, originary from the German low altitude fallow lands, and Shahdara, from the high mountains of Tadjikistan <cite>Loudet02</cite>.<br> | To identify the molecular mechanisms underlying this differences we are performing QTL analysis using a previously developed, well characterized Recombinant Inbred Line set descent from two different natural populations of <i>Arabidopsis thaliana</i>: Bayreuth, originary from the German low altitude fallow lands, and Shahdara, from the high mountains of Tadjikistan <cite>Loudet02</cite>.<br> | ||
We grew replicated individual RILs in environments simulating shade and sun conditions and | We grew replicated individual RILs in environments simulating shade and sun conditions and characterized them on a number of traits associated with the shade avoidance response syndrome. For the QTL analysis we calculated a shade avoidance response index fitting fixed effect models to the phenotipic data, and used an available genetic map for the population that includes more than 500 Single Feature Polymorphism (SFP) markers <cite>West06</cite>.<br> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 33: | Line 43: | ||
<br> | <br> | ||
<br> | <br> | ||
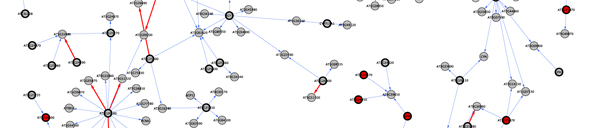
We | We focused in a chromosomal region containing close to 400 genes to fine map and identify the gene responsible for the differences found in the response to shade in the Bay-0 x Sha population. To do this we employed traditional genetic approaches as well as genomic and network analysis. This network analysis is based on coexpression of the candidate genes with other genes across microarray experiments <cite>Riken</cite>, colocalization with expression QTLs <cite>West07</cite>, functional categorization <cite>GO_Classification</cite> and presence of polymorphisms between the parental lines <cite>Clark07</cite>. The use of this bioinformatic approach allowed us to identify ELF3 as the candidate gene for the shade avoidance QTL, which was then confirmed by traditional fine mapping and cloning. | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 43: | Line 53: | ||
|} | |} | ||
<br> | <br> | ||
<h3><font style="color:#F8B603;">Single Nuncleotide Polymorphism discovery | <br> | ||
In the publication of this work, ELF3 alleles of Bay-0 and Sha are shown to differentially affect the shade avoidance response in flowering time and circadian rhtyhms <cite>Jimenez-Gomez10</cite> | |||
<br> | |||
<br> | |||
<h3><font style="color:#F8B603;">Expresion profiling and Single Nuncleotide Polymorphism discovery in cultivated tomato and its wild relatives</font></h3> | |||
---- | ---- | ||
<br> | <br> | ||
Tomato is a specially interesting species because of its natural history, phenotypic diversity among its wild relatives and economic importance. To study the genomic variation among the wild tomato species, we first mined the numerous tomato EST sequences available in the databases in search of polymorphisms. In this dataset, we estimated divergence rates among genes from selected species, and obtained a new set of molecular markers useful in natural variation studies. We performed functional and evolutionary pre-genomic analyses, which gave us an idea of which gene families evolve more rapidly/slowly and have been important during tomato domestication. The results from this work were published <cite>Jimenez-Gomez09</cite> and are available to the community [http://www.plb.ucdavis.edu/labs/maloof/TomatoSNP/index.asp here]. | |||
Now, we are using RNAseq to sequence the transcriptome of four tomato species grown in sun and shade: <i>S. lycopersicum var M82</i>, <i>S. pennellii</i>, <i>S. pimpinellifollium</i> and <i>S. habrochaites</i>. | |||
We developed bioinformatic pipelines to analyze the more than 400 million reads obtained fronm different tissues, species and conditions. The pipeline include scripts that filter and map the reads, detect polymorphisms, calculte their effect on the proteins, perform evolutionary analyses and calculate genome-wide expression levels. Using this methods we identified more than 500.000 polymorphisms in these four speceies and calculated expression differences between species, tissues and environmental conditions. | |||
<br> | <br> | ||
<br> | <br> | ||
| Line 58: | Line 75: | ||
|- | |- | ||
|align="center"|[[Image:PHYB_alignment.jpg|center]] | |align="center"|[[Image:PHYB_alignment.jpg|center]] | ||
<small>amino-acid changes in a fragment of the PHYB gene in 8 | <small>amino-acid changes in a fragment of the PHYB gene in 8 species, red and black bars indicate non-conserved/conserved amino-acid changes respectively</small> | ||
|- | |- | ||
|} | |} | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 78: | Line 88: | ||
#West07 pmid=17179097 | #West07 pmid=17179097 | ||
#Clark07 pmid=17641193 | #Clark07 pmid=17641193 | ||
#Riken [http://prime.psc.riken.jp | #Riken [http://prime.psc.riken.jp Riken] | ||
#GO_Classification pmid=10802651 | #GO_Classification pmid=10802651 | ||
#Jimenez-Gomez09 pmid=19575805 | |||
#Jimenez-Gomez10 pmid=20838594 | |||
</biblio> | </biblio> | ||
|} | |} | ||
__NOTOC__ | __NOTOC__ | ||
Latest revision as of 05:29, 11 January 2012
|
Room 2115 |
QTL and Network analysis of the shade avoidance response in Arabidopsis
Expresion profiling and Single Nuncleotide Polymorphism discovery in cultivated tomato and its wild relatives
Molecular evolution of PHYTOCHROME B
References
|