Frankel:Force Spectroscopy
<owwmenu align="center" font="helvetica" bold="1" color="white" bgcolor="black" hovercolor="black" bghovercolor="orange" topfontsize="10" fontSize="10" image="Danbanner-bio-machines.jpg" >
Home=Frankel
Members=#,Principal Investigator=Frankel:Lab_Members, PhD students=Frankel:Lab_Members, Alumni=Frankel:Lab_Members
Contact=Frankel:Contact
Collaborators=Frankel:Collaborators
Publications=Frankel:Publications
Lab=Frankel:Research
Research=#,Force Spectroscopy=Frankel:Force Spectroscopy,HIV/Virus=Frankel:HIV/Virus,ECM Proteins=Frankel:ECM Proteins,Cyberplasm=Frankel:Cyberplasm,Cancer=Frankel:Cancer
'______ 'Force Spectroscopy
|
Picking up and unfolding a single protein |
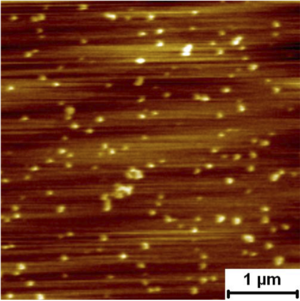
Assembly Nano-mechanics of HIV GP160 GP160mica Typical sawtooth unfolding pattern of a single molecule of the HIV virion protein GP160. Unfolding forces can be extracted from the sawtooth pattern by applying the worm like chain model.
| |
|
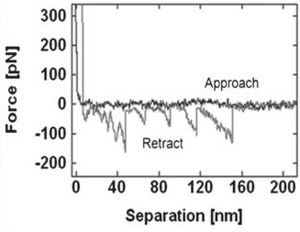
Unfolding the ECM protein fibronectin
Sawtooth pattern on the retraction force curve indicating the unfolding of fibronectin. The average rupture force distribution of the protein on mica surface was 85.1 ± 2.7 pN. |