Drummond:Research: Difference between revisions
Dadrummond (talk | contribs) |
Dadrummond (talk | contribs) (fixes) |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Drummond_Top}} | {{Drummond_Top}} | ||
<div style="width: 750px"> | <div style="width: 750px"> | ||
[[Image:Synthesis-to-misfolding.png]] | |||
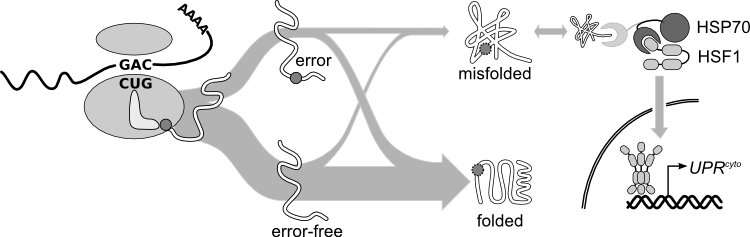
What are the spectrum, frequency and consequences of errors in protein synthesis? How do cells sense and respond to misfolded proteins, particularly in the eukaryotic cytosol? How does stochasticity in protein synthesis alter the composition and stability of the proteome? What benefits do errors confer? Does error-induced protein misfolding influence the progression of neurodegenerative diseases such as ALS? | |||
misfolding | |||
We are pursuing mechanistic answers to these questions, taking a biochemical and genetic approach, with an emphasis on developing high-resolution, high-mass-accuracy mass spectrometric techniques for proteome-scale quantitation. From a theoretical standpoint, we are interested in understanding the imprints that natural selection on fidelity and misfolding leave on evolving genes and genomes. | |||
== | ==Spectrum, frequency, and consequences of errors in protein synthesis== | ||
At the canonical translational misreading error rate of 1 error per 2000 amino acids translated, <em>one in five</em> average-length proteins will contain at least one amino acid not encoded in the messenger RNA. Single amino-acid substitutions frequently destabilize proteins, sometimes disrupting protein folding. Historical measurements of translational error focus on single codons, often in heterologous reporter constructs, leaving open basic questions, such as: how does the frequency of translation errors vary between proteins and along a single protein sequence? Do proteins differ in their tolerance for errors? Are error rates predictable from codons alone, or do other features of transcriptions or nascent polypeptides modulate error rates? | |||
We have assembled evidence that high-expression proteins are under selection to reduce errors (increased translational accuracy), and to fold properly whether error-free or, particularly, despite the errors that do occur (increased translational robustness). We are using high-resolution, high-mass-accuracy mass spectrometry to survey the actual sequences of proteins <i>in vivo</i>, and to address these basic questions about variability in error rates and consequences across the proteome. | |||
< | |||
</ | |||
==Response to misfolded proteins== | |||
When proteins misfold, they become aggregation-prone, toxic species that must be identified and processed by the cell. We study the cytosolic unfolded protein response (UPR<sup>cyto</sup>), the analogue of the well-known unfolded protein response in the endoplasmic reticulum (UPR<sup>ER</sup>). In budding yeast, the appearance of a defined unfolded protein in the cytosol at normal growth temperature induces ~25 interacting proteins involved in cytosolic protein quality control. Taking a biochemical and genetic approach, we are studying the regulation of the UPR<sup>cyto</sup>, asking questions such as: how are unfolded proteins recognized mechanistically? How is this signal propagated into the nucleus to induce gene expression? How does the response return to baseline after the intracellular threat has been dispatched? | |||
<!-- <i>Under construction...thank you for your patience as we remodel to give you greater insight, improved clarity, and more images.</i>--> | |||
{{Drummond_Bottom}} | {{Drummond_Bottom}} | ||
Latest revision as of 19:37, 20 January 2013
What are the spectrum, frequency and consequences of errors in protein synthesis? How do cells sense and respond to misfolded proteins, particularly in the eukaryotic cytosol? How does stochasticity in protein synthesis alter the composition and stability of the proteome? What benefits do errors confer? Does error-induced protein misfolding influence the progression of neurodegenerative diseases such as ALS?
We are pursuing mechanistic answers to these questions, taking a biochemical and genetic approach, with an emphasis on developing high-resolution, high-mass-accuracy mass spectrometric techniques for proteome-scale quantitation. From a theoretical standpoint, we are interested in understanding the imprints that natural selection on fidelity and misfolding leave on evolving genes and genomes.
Spectrum, frequency, and consequences of errors in protein synthesis
At the canonical translational misreading error rate of 1 error per 2000 amino acids translated, one in five average-length proteins will contain at least one amino acid not encoded in the messenger RNA. Single amino-acid substitutions frequently destabilize proteins, sometimes disrupting protein folding. Historical measurements of translational error focus on single codons, often in heterologous reporter constructs, leaving open basic questions, such as: how does the frequency of translation errors vary between proteins and along a single protein sequence? Do proteins differ in their tolerance for errors? Are error rates predictable from codons alone, or do other features of transcriptions or nascent polypeptides modulate error rates?
We have assembled evidence that high-expression proteins are under selection to reduce errors (increased translational accuracy), and to fold properly whether error-free or, particularly, despite the errors that do occur (increased translational robustness). We are using high-resolution, high-mass-accuracy mass spectrometry to survey the actual sequences of proteins in vivo, and to address these basic questions about variability in error rates and consequences across the proteome.
Response to misfolded proteins
When proteins misfold, they become aggregation-prone, toxic species that must be identified and processed by the cell. We study the cytosolic unfolded protein response (UPRcyto), the analogue of the well-known unfolded protein response in the endoplasmic reticulum (UPRER). In budding yeast, the appearance of a defined unfolded protein in the cytosol at normal growth temperature induces ~25 interacting proteins involved in cytosolic protein quality control. Taking a biochemical and genetic approach, we are studying the regulation of the UPRcyto, asking questions such as: how are unfolded proteins recognized mechanistically? How is this signal propagated into the nucleus to induce gene expression? How does the response return to baseline after the intracellular threat has been dispatched?