CH391L/S13/Probiotics: Difference between revisions
No edit summary |
No edit summary |
||
| Line 13: | Line 13: | ||
[[Image:Microbiome_Source2.png|thumb|right|400px]] | [[Image:Microbiome_Source2.png|thumb|right|400px]] | ||
== | == == | ||
(Include paper with E.coli Nissile) | (Include paper with E.coli Nissile) | ||
| Line 23: | Line 23: | ||
*Food Grade Genetic Modification Systems | *Food Grade Genetic Modification Systems | ||
A Food Grade and Generally Recognized as Safe Organism (GRAS) is a classification applied by the US Food and Drug Administration due to its long time use in food products. Most of the Lactococcus lactis strains contain, a GRAS organism, contain multiple plasmids that encode traits important for the food industry. These traits include sugar utilization, bacteriocins, and bacteriophage resistance. Nowadays transfer of between these L. lactis strains occur via the method of bacterial conjugation. A second way to modify L. lactis, less common, is via what we know as [http://openwetware.org/wiki/CH391L/S13/DnaAssembly#Molecular_Cloning molecular cloning] of plasmids between GRAS strains, which has lead to the development of food grade vectors. A methodology used was isolating endogenous plasmids from ''L. lactis'' and separate isolation of useful markers from L.lactis strains that they could then put together into food grade expression Vectors <cite>Dunn2004</cite> | A Food Grade and Generally Recognized as Safe Organism (GRAS) is a classification applied by the US Food and Drug Administration due to its long time use in food products. Most of the ''Lactococcus lactis'' strains contain, a GRAS organism, contain multiple plasmids that encode traits important for the food industry. These traits include sugar utilization, bacteriocins, and bacteriophage resistance. Nowadays transfer of between these ''L. lactis'' strains occur via the method of bacterial conjugation. A second way to modify ''L. lactis'', less common, is via what we know as [http://openwetware.org/wiki/CH391L/S13/DnaAssembly#Molecular_Cloning molecular cloning] of plasmids between GRAS strains, which has lead to the development of food grade vectors. A methodology used was isolating endogenous plasmids from ''L. lactis'' and separate isolation of useful markers from ''L. lactis'' strains that they could then put together into food grade expression Vectors <cite>Dunn2004</cite> | ||
== Oversight of Probiotics == | == Oversight of Probiotics == | ||
Revision as of 11:27, 25 March 2013
Introduction
A probiotic (from the Latin, pro-, "in favor, for" and the Greek , biōtikós, "pertaining to life")refers to live microorganism that provides a benefit to the host, either directly or indirectly, by via interactions with the hosts cells or the host's microbiota. Although research for most of the the last century focused on establishing the fundamentals of what a probiotics is, most of the ongoing research focuses now aims to characterize the microbial communities that have co-evolved with humans, such as Human Microbiome Project that that aims at characterizing the microbial communities found in on the human body and analyzing their roles in our health and disease[1], specific benefits provided by each single organisms to prevent disease. For example, the use of fecal transplantation for antibiotic-associated diarrea [2].(provide actual citation for original paper and summarize.

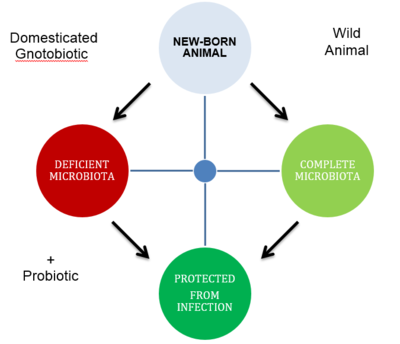
Probiotics
A human's mibrobiota , can be seen as an extended genome. This complex system of microbe-microbe interactions and host-microbe interactions is what allows an state of homeostasis, and when perturbed an state of dysbiosis ensues. Probiotics can alter a host's microbiome to move from the state of dysbiosis to homeostasis. Based on this we can suggest an ideal probiotic would try to achieve reestablishing the benefits in the context of a diet that includes a probiotic either as a supplement or a treatment to a disease. Nowadays this approach can take the form of a rudimentary fecal transplantation from diseased individuals to healthy ones. Their approach to treat antibiotic-associated diarrea caused by Clostridium difficile infection was to assing patient to one of three treatments: 1) included a 4 day w:http://en.wikipedia.org/wiki/Vancomycin vancomycin treatment , (include another example).

(Include paper with E.coli Nissile)
- Another approach has been to genetically modify Streptococcus mutants by deleting lactate dehydrogenase gene and making it defficient in lactic acid production. In turn this same strain became an effector used to produce mutacin which provided an advantage to other strains of S. mutans. This strain was tested in gnobiotic rats was not reported to affect other indigenous flora except for other indigenous S. mutants strains that are associated with dental caries. [3]
Genetic Modification of Probiotics
- Food Grade Genetic Modification Systems
A Food Grade and Generally Recognized as Safe Organism (GRAS) is a classification applied by the US Food and Drug Administration due to its long time use in food products. Most of the Lactococcus lactis strains contain, a GRAS organism, contain multiple plasmids that encode traits important for the food industry. These traits include sugar utilization, bacteriocins, and bacteriophage resistance. Nowadays transfer of between these L. lactis strains occur via the method of bacterial conjugation. A second way to modify L. lactis, less common, is via what we know as molecular cloning of plasmids between GRAS strains, which has lead to the development of food grade vectors. A methodology used was isolating endogenous plasmids from L. lactis and separate isolation of useful markers from L. lactis strains that they could then put together into food grade expression Vectors [4]
Oversight of Probiotics
United States Regulation of Probiotics
Probiotics are currently regulated by the Food and Drug Administration (FDA)in one of the following ways; As a dietary supplement, in which case only a premarket notice to the FDA is necessary or as a drug in which case a premarketing safety, efficacy and approval by the FDA are required. Currently, most of the probiotics on the market fall under the umbrella of a dietary supplement, but situations where the number of infections and the severity of such cases are causing clinicians to evaluate their use as drug, as it's happening for Clostridium difficile infections. In such cases, Florastor (Saccharomyces boulardii) a probiotic currently marketed as a drug is beneficial as it demonstrated its efficacy in reducing the recurrence of C. difficile when used in combination with standard treatment methods. Although cases in which Florastor has lead to fungemia,yeast present in the blood, have been reported, mostly in patients that were not receiving the treatment via introduction of live yeast from contaminated hands of a technician to a catheter site. [5]
iGEM 2009: Stanford's Approach to Probiotics
The 2009 Stanford iGEM Team project centered on probiotics and Inflammatory Bowel Disease (IBD). IBD, as explained, is caused by an imbalance of two types of T-cells, Treg cells that immunosuppres the Th17 cells that cause the inflammation seen in patients. They suggest that an novel theraputic mechanism can be achieved by in vivo regulation of these cells. Their approach focuses in constructing two different Escherichia coli(E.coli) strains, each that would contain a distinct input/output cassette , each that is referred as a device. The first device would detect as input Nitric Oxide(NO), a byproduct of inflammation and Th17 proliferation, produces retinoic acid, that blocks further CD4+ T-cells differentiation into Th17 cells. The second device detects 5-Methyl tryptophan (5MT) as an input and produces Interleukin-6 to regulate Treg proliferation to regulate their immunosuppression response. Ideally depending on the balance between these two markers, if too much NO is sensed by Device 1 it would prevent inflammation. The opposite would also be true if the second device sences to much 5MT that would immunosuppress Th17 cell by blocking their differentiation.

Probiotics and the Media links
The Media perspective on Probiotics
- http://www.npr.org/2011/09/02/140146780/probiotic-bacteria-chill-out-anxious-mice
- http://www.telegraph.co.uk/health/healthnews/8261808/Designer-probiotic-yogurts-could-help-people-lose-weight.html
References
- Gordon JI. Honor thy gut symbionts redux. Science. 2012 Jun 8;336(6086):1251-3. DOI:10.1126/science.1224686 |
- Borody TJ, Warren EF, Leis SM, Surace R, Ashman O, and Siarakas S. Bacteriotherapy using fecal flora: toying with human motions. J Clin Gastroenterol. 2004 Jul;38(6):475-83. DOI:10.1097/01.mcg.0000128988.13808.dc |
- Hillman JD. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):361-6.
- Liu CQ, Su P, Khunajakr N, Deng YM, Sumual S, Kim WS, Tandianus JE, and Dunn NW. Development of food-grade cloning and expression vectors for Lactococcus lactis. J Appl Microbiol. 2005;98(1):127-35. DOI:10.1111/j.1365-2672.2004.02441.x |
- Venugopalan V, Shriner KA, and Wong-Beringer A. Regulatory oversight and safety of probiotic use. Emerg Infect Dis. 2010 Nov;16(11):1661-5. DOI:10.3201/eid1611.100574 |
- Kinross JM, Darzi AW, and Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011 Mar 4;3(3):14. DOI:10.1186/gm228 |
- Bengmark S. Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut. 1998 Jan;42(1):2-7. DOI:10.1136/gut.42.1.2 |
-
pmid=