CH391L/S13/In vitro Selection of FNAs: Difference between revisions
No edit summary |
No edit summary |
||
| Line 18: | Line 18: | ||
===Deoxyribozymes=== | ===Deoxyribozymes=== | ||
[[Image:8-17OP.png]] | [[Image:8-17OP.png]] | ||
===Aptamers and Riboswitches=== | |||
Revision as of 02:26, 11 February 2013
Introduction
Functional nucleic acids (FNAs) are RNA, DNA, or XNA(nucleic acid analogues) that perform an activity such as binding or catalyzing a reaction. FNAs are grouped into three main categories Aptamers, Ribozymes, and Deoxyribozymes that are subdivided into either natural or artificial depending on their origin; the exception being Deoxyribozymes as they have yet to be discovered in a living organism.
Functional Nucleic Acids
Ribozymes
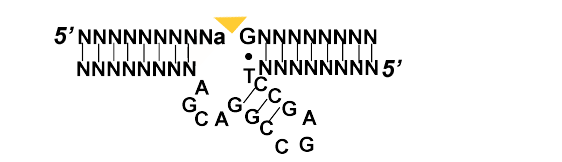
Deoxyribozymes
Aptamers and Riboswitches
In vitro Selection of Functional Nucleic Acids
Extra
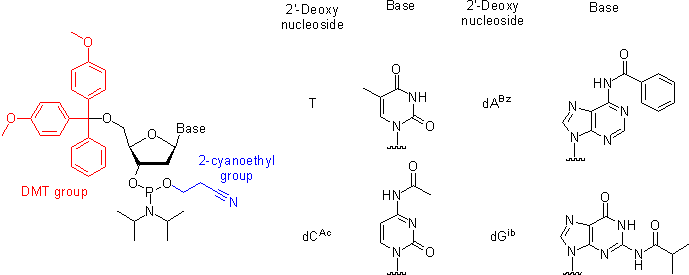
Oligonucleotides are chemically synthesized from DNA phosphoramidite monomers. Briefly, activated phosphoramidite monomers are added in the 3' to 5' direction using a cyclical activation and blocking chemistry to obtain a DNA polymer linked by phosphodiester bonds.
Chemical synthesis is currently limited to oligonucleotides of about 200 nt in length. <biblio>
- Cech1982 pmid=6297745
- Altman1983 pmid=6197186
<\biblio>