CH391L/S13/In vitro Selection of FNAs: Difference between revisions
No edit summary |
No edit summary |
||
| Line 12: | Line 12: | ||
<cite>Cech1982</cite>,<cite>Altman1982</cite> | <cite>Cech1982</cite>,<cite>Altman1982</cite> | ||
== | |||
===Ribozymes=== | |||
===Deoxyribozymes=== | |||
== | ==In vitro Selection of Functional Nucleic Acids== | ||
[[Image:Image:In-vitro-selection.png]] | |||
Revision as of 00:53, 11 February 2013
Introduction
Functional nucleic acids (FNAs) are RNA, DNA, or XNA(nucleic acid analogues) that perform an activity such as binding or catalyzing a reaction. FNAs are grouped into three main categories Aptamers, Ribozymes, and Deoxyribozymes that are subdivided into either natural or artificial depending on their origin; the exception being Deoxyribozymes as they have yet to be discovered in a living organism.
Functional Nucleic Acids
Ribozymes
Deoxyribozymes
In vitro Selection of Functional Nucleic Acids
File:Image:In-vitro-selection.png
Extra
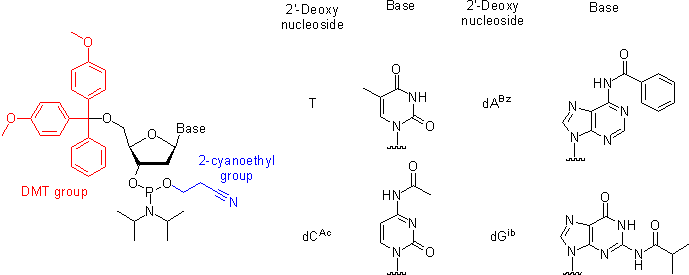
Oligonucleotides are chemically synthesized from DNA phosphoramidite monomers. Briefly, activated phosphoramidite monomers are added in the 3' to 5' direction using a cyclical activation and blocking chemistry to obtain a DNA polymer linked by phosphodiester bonds.
Chemical synthesis is currently limited to oligonucleotides of about 200 nt in length. <biblio>
- Cech1982 pmid=6297745
- Altman1983 pmid=6197186
<\biblio>