CH391L/S13/In vitro Selection of FNAs: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
(New page: Category:CH391L_S13 ==Introduction== ==Types of Functional Nucleic Acids== <!--Regardless of the length of the eventual product, synthetic DNA constructs are built from some co...) |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Category:CH391L_S13]] | [[Category:CH391L_S13]] | ||
==Introduction== | |||
==Types of Functional Nucleic Acids== | |||
== | ==In vitro Selection of Functional Nucleic Acids== | ||
==Ribozymes== | |||
==Deoxyribozymes== | |||
==Aptamers== | |||
== | ==Extra== | ||
<!--Regardless of the length of the eventual product, synthetic DNA constructs are built from some combination of short DNA oligonucleotides. These oligonucleotides are later assembled into a complementary DNA duplex, amplified and inserted into their final genetic context. | <!--Regardless of the length of the eventual product, synthetic DNA constructs are built from some combination of short DNA oligonucleotides. These oligonucleotides are later assembled into a complementary DNA duplex, amplified and inserted into their final genetic context. | ||
| Line 19: | Line 30: | ||
Chemical synthesis is currently limited to oligonucleotides of about 200 nt in length. | Chemical synthesis is currently limited to oligonucleotides of about 200 nt in length. | ||
Revision as of 15:43, 8 February 2013
Introduction
Types of Functional Nucleic Acids
In vitro Selection of Functional Nucleic Acids
Ribozymes
Deoxyribozymes
Aptamers
Extra
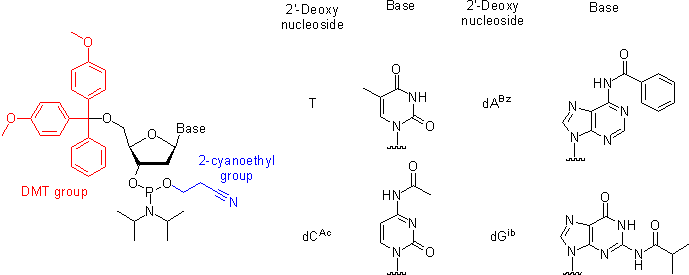
Oligonucleotides are chemically synthesized from DNA phosphoramidite monomers. Briefly, activated phosphoramidite monomers are added in the 3' to 5' direction using a cyclical activation and blocking chemistry to obtain a DNA polymer linked by phosphodiester bonds.
Chemical synthesis is currently limited to oligonucleotides of about 200 nt in length.