CH391L/S13/Biocontainment
Biocontainment
Biocontainment is a set of strategies to confine pathogenic and genetically modified organisms to the laboratory. Biocontainment is a concern in medical, agricultural, and synthetic biology operations. With many companies performing synthetic biology research and production biocontainment is of increased concern, as more and more exotic GMO’s are produced. Presently, there are no differences between biocontainment strategies for GMOs than there are for synthetic biology. Typicaly biocontainment is a mixture of physical barriers as well as an emerging field of genetic barriers placed on an organism to prevent it from entering the environment. Physical barriers as part of a biocontainment strategy are a part of general lab biosafety practices (http://en.wikipedia.org/wiki/Biocontainment).
In 1993 the NIH produced a set of guidelines for research involving recombinant DNA molecules, in the report it is recommended that engineered microbe survival or engineered DNA transmission is less than 1 cell per 10^8 cells (Wilson,1993). Currently, many genetic barrier strategies are able to reduce genetic transmission or provide induced lethality within these guidelines. There have been observed a great number of instances of GMOs escaping from the lab (Haynes 2013). However, there have been no bio-hazardous accidents linked to either GMOs or synthetic biology (Schmidt and de Lorenzo, 2012).
History of biocontainment for GMOs
The first use of a genetic barrier for a strategy of biocontainment was reported in 1987. The hok gene (encoding for a lethal toxin) was incorporated on a plasmid, this strategy prevents the organism from being viable when leaving the lab, and the plasmid cannot be incorporated in to other organisms, as it is encoding the toxin gene. (Molin et al., 1987)
Containment strategies
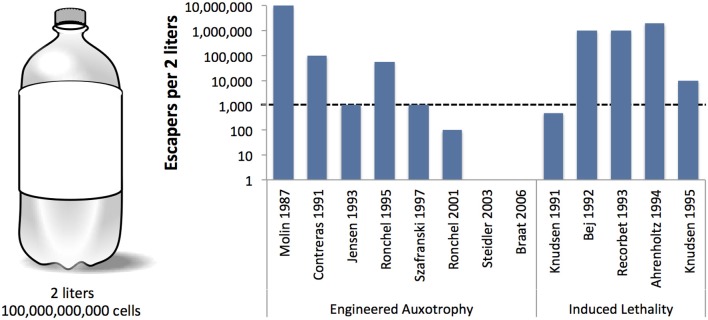
Engineered Auxotrophy
Auxotrophy is the inability of an organism to synthesize a particular organic compound required for its growth. In synthetic biology this is used to create an organism that cannot grow when it leaves the lab or a production facility, because it would be unlikely to encounter the necessary metabolite in nature. This has been a strategy in research for over 70 years, made famous in Beadle and Tatum's Nobel prize-winning work on the one gene-one enzyme hypothesis.
Examples of engineered auxotrophy
1991 Conditional-Suicide Containment System for Bacteria Which Mineralize Aromatics – Here they have pseudoimonas that degrades benzoates for bioremediation, and then dies when substrate is no longer present. ( Contreras et al. (1991) 2003 Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10 – Here they have engineered a Lactococcus that produces il-10 for the treatment of Crohn’s disease, this strain requires thymidine for full viability as it has a knockout of thyA. (Steidler et al., 2003)
Induced Lethality
Is a biocontainment strategy in which GMOs are contained by the induction of toxic genes upon leaving the lab setting. The toxin/antitoxin systems of bacteria have been used in many papers examining the use of induced lethality. This induction is typically controlled by the presence or absence of an induction molecule, but theoretically any inducible promoter can be adapted to this purpose. More recently, the idea of induced lethality that has been engineered to cause death after a certain amount of cell cycles has been proposed (Friedland et al., 2009).
Examples of Induced Lethality
1991 - Development of efficient suicide mechanisms for biological containment of bacteria. – They use a lactose inducible system for the production of the RelF (Knudsen and Karlström, 1991).
1994 - A conditional suicide system in Escherichia coli based on the intracellular degradation of DNA. – They used a heat inducible system paired with a nuclease gene (Ahrenholtz et al., 1994)
Gene-Flow barriers
Gene-flow barriers are required to prevent the transmission of genetic material from the original host organisms to organisms outside of the lab. These systems typically have a toxin gene or nuclease that the host is immune to, but when the transgenic DNA enters another host organism it will result in death. In plants, recombinant DNA can be sequestered in the chloroplasts resulting in another physical barrier. DNA integrated in chromosomes should have a lower likelihood of transmission between species than those contained on plasmids.
Example of gene-flow barriers
2003 - A dual lethal system to enhance containment of recombinant micro-organisms. - In this paper they had the gene with the trait they wanted flanked by two toxin antitoxin systems. (Torres, 2003).
Orthogonal genetics
Orthogonal genetic strategies for biocontainment (semantic containment) strategies include using XNAs for encoding desired protein or rearranging codons between canonical amino acids. This is an active area of research in synthetic biology but no real world biocontainment strategy using these methods has been deployed as of yet.
Concerns about genetic biocontainment strategies
Genetic safeguards for preventing organisms from entering the environment are subject to spontaneous mutations and can cause failure to many biocontainment strategies. Many strategies are developing that contain multiple genetic barriers.
Outreach about Synthetic Biology and Biocontainment
The Woodrow Wilson Synthetic Biology Project was developed to establish a public and policy discourse about synthetic biology. The project makes effort to discuss biosafety involved with synthetic biology. They recently published a paper on biosafety and synthetic biology and what they believe needs to be further researched (Dana et al., 2012).
The four steps from the paper:
- 1) How can synthetic organisms physiology disrupt nature?
- 2) How can synthetic organisms disrupt habitats?
- 3) Can synthetic organism evolve quickly and persist in habitats?
- 4) How much risk is associated with gene transfer?
IGEM Connection
The 2012 UCSF IGEM team used a toxin/antitoxin system to tune strain ratios in E. coli. The project overview is described here: http://2012.igem.org/Team:UCSF/Toxin_System and the project results are described here: http://2012.igem.org/Team:UCSF/Toxin_Data
References
<biblio>
- Haynes2013 pmid=23355834
- Lorenzo2012 pmid=22710182
- Dana2012 pmid=22382962

