Biomolecular Breadboards:Preliminary Data: Difference between revisions
Clare Hayes (talk | contribs) mNo edit summary |
|||
| Line 7: | Line 7: | ||
Expression of plasmids can be optimized by concentration. | Expression of plasmids can be optimized by concentration. | ||
[[Image: | [[Image:plasmid_sat.png|400px]] | ||
===Protecting Linear DNA from Exonuclease-Mediated Degradation=== | ===Protecting Linear DNA from Exonuclease-Mediated Degradation=== | ||
Current standards for circuit design utilize plasmids for DNA template, which require time-consuming subcloning steps. However, circuits based on linear DNA require only PCR assembly or gene synthesis, which drastically decreases preparation time. As a purely extract-derived system, our biomolecular breadboard exhibits exonuclease activity which degrades linear DNA. | Current standards for circuit design utilize plasmids for DNA template, which require time-consuming subcloning steps. However, circuits based on linear DNA require only PCR assembly or gene synthesis, which drastically decreases preparation time. As a purely extract-derived system, our biomolecular breadboard exhibits exonuclease activity which degrades linear DNA. We are developing multiple technologies to protect linear DNA from exonuclease degradation. | ||
We are developing multiple technologies to protect linear DNA from exonuclease degradation. | |||
====Protection Sequences==== | ====Protection Sequences==== | ||
| Line 23: | Line 20: | ||
====GamS==== | ====GamS==== | ||
[[Image:gamSComp.png|400px]] | [[Image:gamSComp.png|400px]] [[Image:linear_sat.png|right|400px]] | ||
One main 3’ exonuclease present is the RecBCD complex, which can be inhibited by gamS protein produced by lambda phage. | One main 3’ exonuclease present is the RecBCD complex, which can be inhibited by gamS protein produced by lambda phage. | ||
Revision as of 23:42, 11 July 2012
| Home | Protocols | DNA parts | Preliminary Data | Models | More Info |
Preliminary Data
Plasmid Expression of GFP
Using pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500, a plasmid enhanced for GFP expression, the biomolecular breadboard is able to express mass at equal concentration to comparable bacteriophage in-vitro systems (J. Shin and V. Noireaux, 2010).
Expression of plasmids can be optimized by concentration.
Protecting Linear DNA from Exonuclease-Mediated Degradation
Current standards for circuit design utilize plasmids for DNA template, which require time-consuming subcloning steps. However, circuits based on linear DNA require only PCR assembly or gene synthesis, which drastically decreases preparation time. As a purely extract-derived system, our biomolecular breadboard exhibits exonuclease activity which degrades linear DNA. We are developing multiple technologies to protect linear DNA from exonuclease degradation.
Protection Sequences
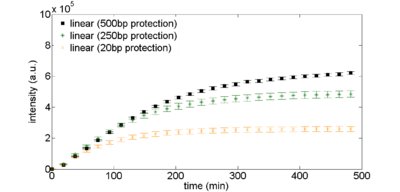
Figure 1: Exonuclease protection using non-coding DNA. Linear DNA templates, 2nM, are derived from pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 with varying amounts of noncoding DNA surrounding the coding sequence.
GamS

One main 3’ exonuclease present is the RecBCD complex, which can be inhibited by gamS protein produced by lambda phage.


