BISC219/F12: Lab 6
LAB 6 Mapping Continued: Making the males to be able to set up a test cross
You will transfer 3-4 L4 hermaphrodite true breeding double mutants to 2 separate cross plates. Then add 3-4 wild type males to each plate in order to create male heterozygotes (++/ d u).
To make sure you have fertile L4 double mutant hermaphrodites for the test cross that you will set up in 3 days, transfer a single double mutant from your true breeding plate to each of 2 separate maintenance plates.
Label the plates you will set up today with your PURPLE Sharpie. Incubate your worms at 23C for 3 days.
We will use the double mutants and these male heterozygotes to perform a test cross in 3 days. This test cross is designed in such a way that we can differentiate the progeny of parental gametes from those of recombinant gametes PHENOTYPICALLY. Review the mapping project crosses that you diagrammed for homework and make sure you understand how and why our test cross allows this crucial differentiation. We will score the progeny of that test cross as either parental (WT or double mutants) or recombinant (Unc or Dpy single mutants) and use the Recombination Frequency (number of single mutants) to calculate map distance of our dpy gene of interest from the linked unc. This scoring and calculation will occur in the next lab.
To do in 3 days
FINALLY!!! Set up the crucial TEST CROSS: Return to lab and transfer 3-5 L4 double mutant hermaphrodites to each of 2 crossing plates. You will find the clean crossing plates in your lab day's box in the supply area in the back of the lab. Add 3-5 of the heterozygous males (phenotypically wild type) of genotype (++/ d u) that you created in your last cross to each of those cross plates with the double mutant hermaphrodites. Label these plates with your PURPLE Sharpie.Incubate all worms at 23C until your next lab period.
Forward Genetics: Sequencing Analysis
Once we know our gene's name or its mapped location on a chromosome we can design primers that will allow is to amplify the gene of interest by polymerase chain reaction and then use those templates in chain termination sequencing reactions (Sanger sequencing) to provide us with direct evidence of the nature of the mutation, specifically the base alteration in the mutants in comparison to the wild type sequence. Understanding the nature and extent of the change in the gene may allow us to pinpoint a functionally significant part of the gene product (usually a protein). Understanding the nature of the change in the polypeptide sequence of the protein is useful in understanding how gene products function in organisms. For example, if a single point mutation causes an amino acid to change from a hydrophobic to a hydrophilic R group, or a flexible to an inflexible amino acid, the entire protein could misfold. As you know, shape is crucially important to function in enzymes, in DNA binding proteins, and in other proteins, all of which can have important regulatory functions in the organism.
Chain Termination DNA Sequencing (also called Sanger sequencing):
There is a nice animation of this process done by the Dolan DNA center at: | http://www.dnalc.org/resources/animations/sangerseq.html.
There are three major steps in a sequencing reaction, which are repeated for 25 cycles in a thermocycler:
- Denaturation at 94°C: During the denaturation, the double strand melts open to single stranded DNA and all enzymatic reactions stop, including the extension from the previous cycle.
- Annealing at 50°C: In each sequencing reaction, a specific primer is required. This primer is a short sequence of bases complementary to a region of the plasmid upstream of the target gene. The primer will anneal and stay, we hope, only at one position on the single stranded DNA template. In sequencing reactions there is only one strand copied whereas, in PCR, two primers are used so both strands of the DNA template are copied. The primer jiggles around, caused by the Brownian motion. Ionic bonds are constantly formed and broken between the single stranded primer and the single stranded template. The more stable bonds last a little bit longer (primers that fit exactly) and on that small section of double stranded DNA (template and primer), the polymerase can attach and starts copying the template. Once there are a few bases built in, the ionic bond is so strong between the template and the primer, that it does not break anymore.
- Extension at 60°C: Sixty degrees is the ideal working temperature for polymerase activity in sequencing even though, normally, it is 72 °C. Because the growing strand must incorporate ddNTP's which are chemically modified with a fluorescent label, the temperature is lowered so it has time to incorporate the 'strange' molecules. Well designed primers, where there are a few bases built in, already have a stronger ionic attraction to the template than the forces breaking these attractions. Primers that are on positions with no exact match, come loose again and don't give an extension of the fragment. The bases (complementary to the template) are coupled to the primer on the 3'side (adding dNTP's or ddNTP's from 5' to 3', reading from the template from 3' to 5' side, bases are added complementary to the template). When a ddNTP is incorporated, the extension reaction stops because a ddNTP contains a H-atom on the 3rd carbon atom (dNTP's contain a OH-atom on that position). Since the ddNTP's are fluorescently labeled, it is possible to detect the color of the last base of this fragment on an automated sequencer.
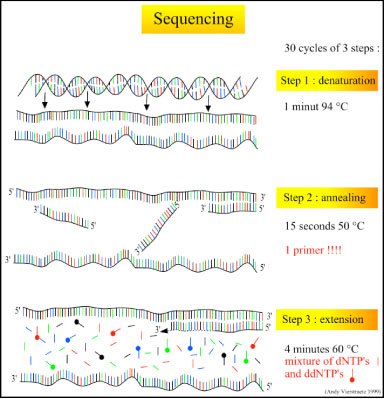
Because only one primer is used in sequencing reactions, only one strand is copied, and there is a linear (not log as in PCR) increase of the number of copies of one strand of the gene. Therefore, there has to be a large number of copies of the gene in the starting mixture for optimal sequencing. If there are 1000 copies of the wanted gene before the cycling starts, after one cycle there will be 2000 copies: the 1000 original templates and 1000 complementary strands with each one fluorescent label on the last base. After two cycles, there will be 2000 complementary strands, three cycles will result in 3000 complementary strands and so on. Linear amplification during sequencing reactions explains why we have to start with an exponential amplification of the gene of interest in a polymerase chain reaction to get lots of copies of our gene BEFORE we do the sequencing. Both sequencing and polymerase chain reactions require short sequences of DNA called primers to direct the specificity of the amplification to only the gene of interest or a part of the gene of interest. Primers must be designed to anneal to a short section of DNA on each strand adjacent to the section of the genome you want to copy. This means that you MUST know the sequence of the DNA near or of your gene of interest. Fortunately the entire C. elegans genome has been sequenced; therefore we can use Wormbase to locate where we want to start sequencing (by gene name or map location), find the sequence of the wild type form of the gene, and design primers to anneal at the proper places and to direct the specificity of the sequencing of our mutant gene. Note that you needed to complete either the mapping or the complementation analysis before DNA sequencing of the mutated gene so that we could look up its sequence in Wormbase and design and order short sequences of DNA that will anneal and copy that gene only or a section of that gene.

What your instructor did for you:
Due to time restraints, your instructor did the preparation of the worm DNA for sequencing (digestion of the Dpy worm to release the DNA & amplification and purification of the gene of interest by PCR with primers designed from the Wormbase sequence of the gene of interest). She then sent away the purified PCR product (amplified mutant gene) and the same primers to a commercial sequencing facility where they did chain termination (Sanger) sequencing reactions from those primers and our template DNA (the PCR product). They then used an automatic sequencer to separate the fluorescently labeled gene fragments by size, from smallest to largest numbers of bases, and a laser read the colors on the end of each fragment. A computer turned that information into .abi or .seq files. Those files of the mutant sequence, along with reference files of the wild type version of the gene, are posted to a folder on the Desktop of the lab computers. They are now available for you to analyze in your effort to functionally characterize the gene associated with the Dpy phenotype with which we began.
Steps in sequencing a C. elegans gene:
1. Digest the tough outer cuticle.
Add worms to a mixture of 10 mM Tris, 50 mM KCl, 1.5 mM MgCl2, pH 8.3, with 10 mg/ml Proteinase K.
Freeze the worms at -80°C for 15 minutes
Incubate at 65°C for 1 hour to digest the cuticle and then at 95°C for 15 minutes to denature the Proteinase K.
2. Amplify the gene of interest by Polymerase Chain Reaction (PCR) using primers designed to amplify only this gene or a part of this gene.
Add dNTPs to a final concentration of 0.2 mM and primers to a final concentration of 0.4 mM each.
Add Taq according to manufacturer's instructions.
Determine the proper reaction conditions for your primers and gene size.
After amplification check for success via agarose gel electrophoresis.
3. Purify the product and send for sequencing
Remove all impurities and chemicals from the PCR product using a Qiagen PCR Purification Kit
following manufacturer's instructions.
Quantify DNA concentration.
Send proper concentration of DNA and primers to company for sequencing. Primers were designed from looking up the gene of interest (or ORF region)'s sequence in WormBase and figuring out complementary short sequences at the beginning and end of the region we want to sequence. We have used Genewizas our commercial sequencing facility.
What you need to do:
To analyze your sequences, you will need to work with software that can read .abi and .seq files that are generated by automatic sequencers. If there is time, your instructor will show you our ABI automatic sequencer and explain its operation. The directions for using the DNAstar software by Lasergene to analyze your sequences are found at: Media:Instructions for DNA sequence analysis F12_2.doc
Assignment
Remember to check the Assignment section of the wiki for instructions about the graded assignment due in the next lab and check the Weekly Planner section of the Calendar/Lab Planner for other work to accomplish before the next lab. Direct link to information on the written assignment due in Lab 7 is at Assignment Mapping Crosses. BISC219/F12: Assignment_Series2_DNA Sequencing.
