BIO254:Exam2002: Difference between revisions
| Line 45: | Line 45: | ||
(c) Wow! Your mutant has a striking phenotype. You notice that in a subpopulation of neurons your mutant axons fail to make a critical turn at a specific point (neuron A). You are not sure if your newly discovered receptor acts as a receptor for an attractive or repulsive cue. Draw two models postulating an attractive or repulsive cue. (6 pts) | (c) Wow! Your mutant has a striking phenotype. You notice that in a subpopulation of neurons your mutant axons fail to make a critical turn at a specific point (neuron A). You are not sure if your newly discovered receptor acts as a receptor for an attractive or repulsive cue. Draw two models postulating an attractive or repulsive cue. (6 pts) | ||
[[Image:Exam2002Pic5. | [[Image:Exam2002Pic5.jpg]] | ||
| Line 66: | Line 66: | ||
[[Image:Exam2002Pic4. | [[Image:Exam2002Pic4.jpg]] | ||
Wild-type Wild-type | Wild-type Wild-type | ||
Revision as of 15:53, 3 December 2006
Bio154 Sample Exam 2002-Key
NOTE: Answers are provided by your classmates. Some questions may have multiple correct answers; as long as yours is reasonable, logic and consistent, you can get full credit. Some questions may be challenging but don’t panic; grading will be on a curve. Good luck.
I. (12 pts)
Describe three major similarities and three major differences (biology and/or mechanism) of Rac (a Rho family small GTPase) and Transducin (a trimeric G protein) (The fact that they are both GTPases is a given now.)
II. (24 pts)
(a) While searching the human genome you come across a putative receptor for axon guidance based on its conserved motifs with other known axon guidance receptors. You are happy to find out that this receptor has unstudied homologues in other organisms including worm, fly, zebra fish, and mouse. Give two reasons why you should be happy about that (4 pts).
(b) You decide to make a loss-of-function mutant of this putative receptor in fly. How will you determine where to look in your mutant animal for phenotypes? (2 pts) Give a reason why you might not see a phenotype even if you have “knocked out” the gene activity completely (2 pts).
(c) Wow! Your mutant has a striking phenotype. You notice that in a subpopulation of neurons your mutant axons fail to make a critical turn at a specific point (neuron A). You are not sure if your newly discovered receptor acts as a receptor for an attractive or repulsive cue. Draw two models postulating an attractive or repulsive cue. (6 pts)
Wild-type Mutant
(d) You also notice that in wild-type animals another subpopulation of neurons never make this turn (neuron B). Propose an experiment to show that your receptor is sufficient for axon turning in this system. (6 pts)
Wild-type Wild-type
(e) You are now interested in what is downstream of your receptor. Propose two general experimental techniques that may help you answer this question. (4 pts)
III. (24 pts)
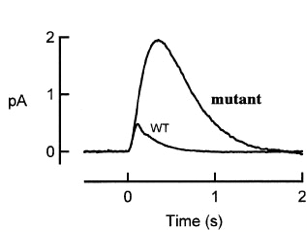
An interesting mutant of gene X has been found in the course of photoreceptor study. The figure below shows the population mean single photon responses of rods from wild type and from mutant X.
1) According to the picture above, what step of the photoreceptor response is affected by X mutant? (4 pts)
2) What protein could gene X encode? Give three candidates discussed in lectures. Explain how mutations in these three candidate genes could result in the phenotypes seen in the graph. (12 pts)
3) Come up with two further experiments to distinguish your 3 hypotheses in (2). (8 pts)
IV. (24 pts)
To investigate the mechanisms of retinocollicular (mammalian equivalent of retinotectal) projection specificity, O’Leary and colleagues generated knock-out mice for both Ephrin B2 and B3 receptors (EphB2 & EphB3). To study the phenotype, they inject an axon tracer DiI into a very small and identical region of the retina in both WT and double knockout mice, and then analyze where these RGC axons terminate in the superior colliculus by following DiI labeled axons. The figure shows the DiI labeled RGC axon termination in the superior colliculus for WT and double knockout mice (L: lateral; M: medial; P: posterior; Anterior border is indicated by the arrowheads).
...............WT....................EphB2-/-;EphB3-/-.....
1) Judging from the location of the termination in the superior colliculus in WT, which part of the retina do you think they inject their DiI? (4 pts)
2) In class, we mentioned that EphBs are expressed in RGC in a high ventral-low dorsal gradient; and Ephrin Bs are expressed in superior colliculus in a high medial-low lateral gradient. How can you explain the double knock-out phenotype? (4 pts)
3) If EphB2 and EphB3 are the only receptors for D-V patterning, you would imagine that without these receptors, axons will randomly distribute along the D-V axis. However you quickly notice that the ectopic terminal zones (eTZ) always project lateral to the correct terminal zone (TZ) instead diffusely in both directions (see Figure). Propose a simple model that explains this effect. (4 pts)
4) You may also have noticed that in EphB mutants axons in eTZ largely concentrate on one spot instead of being diffusive. Propose an explanation for this phenomenon and describe a simple experiment to test your hypothesis. (6 pts).
5) It turns out that Ephrin Bs are also expressed in RGC axons in a gradient fashion along D-V axis, and EphB are also expressed in superior colliculus (target) along a M-L axis! Moreover, since the “ligands” for EphBs, the Ephrin B, are transmembrane proteins so they can sometimes act as receptors for “reverse signaling”. So the phenotypes seen in the Figure is not necessarily caused by loss of EphBs in the axons; the loss of EphB in the target could also contribute. Describe one experiment that could distinguish whether EphB is needed in RGC axon or in the target for the correct establishment of topographic map along the D-V axis. (6 pts)
V. (16 pts)
1) What is the difference between voltage-dependent closure of a channel and inactivation of a channel? (3 pts)
2) Describe one example in which inactivation independent of voltage-dependent closure is important. (3 pts)
3) Describe one mechanism by which inactivation is achieved. (2 pts)
4) Type-A potassium channels are a group of voltage-gated K channels that rapidly inactivate at membrane potentials at which the classic Hodgkin-Huxley voltage dependent potassium channel stays active.
In the voltage-clamp experiment shown below, a molluscan neuron containing classic Hodgkin-Huxley K channels and type-A potassium channels was depolarized to 5 mV from a holding potential of either -40 mV or -80 mV. The resulting current is quite different: one (from -40mV) consists of only IK (the classic Hodgkin-Huxley channel), the other (from –80mV) consists of IK and IA (the current from the type A channel). One could thus deduce the IA current from subtraction (dotted line).

(a) What does this experiment reveal about the properties of the type-A channel? (4 pts)
(b) Type-A potassium channels prevent neurons from firing repetitively at high frequencies but enable them to fire repetitively at low frequencies. How do the properties of the type-A channel enable it to regulate the spacing of action potentials? (4 pts)
Recent updates to the site:
- N
- This edit created a new page (also see list of new pages)
- m
- This is a minor edit
- b
- This edit was performed by a bot
- (±123)
- The page size changed by this number of bytes
24 April 2024
|
|
08:14 | "Pick and Place" Assembly of Parts Using PDMS - Amy Lim, Rylie Costello 6 changes history +394 [Rcostello (6×)] | |||
|
|
08:14 (cur | prev) +1 Rcostello talk contribs (→"Pick and Place" Methodology) | ||||
|
|
08:13 (cur | prev) −14 Rcostello talk contribs (→"Pick and Place" Methodology) | ||||
|
|
08:12 (cur | prev) −1 Rcostello talk contribs (→"Pick and Place" Methodology) | ||||
|
|
08:12 (cur | prev) −1 Rcostello talk contribs (→"Pick and Place" Methodology) | ||||
|
|
08:12 (cur | prev) +110 Rcostello talk contribs (→References) | ||||
|
|
08:11 (cur | prev) +299 Rcostello talk contribs (→"Pick and Place" Methodology) | ||||
| 08:02 | Upload log Rcostello talk contribs uploaded File:Pick and Place.mp4 | ||||
|
|
06:23 | WAKNA:Basics 2 changes history −229 [Berthold Drexler (2×)] | |||
|
|
06:23 (cur | prev) +192 Berthold Drexler talk contribs (→Hier finden Sie Literatur für Einsteiger:innen in das Gebiet der Neuroanästhesie) | ||||
|
|
06:21 (cur | prev) −421 Berthold Drexler talk contribs (→Sonstige) | ||||
23 April 2024
|
|
15:33 | "Pick and Place" Assembly of Parts Using PDMS - Amy Lim, Rylie Costello 6 changes history +837 [Rcostello (6×)] | |||
|
|
15:33 (cur | prev) +1 Rcostello talk contribs (→"Pick and Place" for Microfluidics) | ||||
|
|
15:33 (cur | prev) +203 Rcostello talk contribs (→References) | ||||
|
|
15:31 (cur | prev) −2 Rcostello talk contribs (→"Pick and Place" for Microfluidics) | ||||
|
|
15:29 (cur | prev) −474 Rcostello talk contribs (→References) | ||||
|
|
15:29 (cur | prev) +845 Rcostello talk contribs (→MEMS Devices) | ||||
|
|
15:14 (cur | prev) +264 Rcostello talk contribs (→"Pick and Place" for Microfluidics) | ||||
|
|
11:58 | BioMicroCenter:People 2 changes history +30 [Lttran (2×)] | |||
|
|
11:58 (cur | prev) −4 Lttran talk contribs (→BioMicro Center Staff) | ||||
|
|
11:49 (cur | prev) +34 Lttran talk contribs (→BioMicro Center Staff) | ||||
| 11:46 | Upload log Lttran talk contribs uploaded File:SKR BMC.jpg | ||||
22 April 2024
|
|
19:28 | "Pick and Place" Assembly of Parts Using PDMS - Amy Lim, Rylie Costello 4 changes history +1 [Rcostello (4×)] | |||
|
|
19:28 (cur | prev) −2 Rcostello talk contribs (→Nanowires) | ||||
|
|
19:26 (cur | prev) 0 Rcostello talk contribs (→Biology-Inspired Solution) | ||||
|
|
15:03 (cur | prev) +2 Rcostello talk contribs (→At the Microscale) | ||||
|
|
15:02 (cur | prev) +1 Rcostello talk contribs (→Overview) | ||||
| 09:24 | CHEM-ENG590E:Wiki Textbook diffhist +16 Rcostello talk contribs (→Chapter 15 - Other Topics) | ||||
| 09:24 | Move log Rcostello talk contribs moved page "Pick and Place" Assembly of Parts Using PDMS - Amy Lim to "Pick and Place" Assembly of Parts Using PDMS - Amy Lim, Rylie Costello | ||||
| 08:59 | "Pick and Place" Assembly of Parts Using PDMS - Amy Lim diffhist −2,792 Rcostello talk contribs (→"Pick and Place" for Microfluidics) | ||||
21 April 2024
| 22:55 | Lee:JC diffhist +24 Wooin Lee talk contribs (→2024) | ||||
| 17:42 | Ernesto-Perez-Rueda:Publications diffhist +55 Ernesto Perez-Rueda talk contribs | ||||
19 April 2024
|
|
21:58 | Hu 2 changes history +58 [Hugangqing (2×)] | |||
|
|
21:58 (cur | prev) −8 Hugangqing talk contribs | ||||
|
|
21:58 (cur | prev) +66 Hugangqing talk contribs | ||||