IGEM:Imperial/2010/Modelling: Difference between revisions
Anita Nguyen (talk | contribs) |
Anita Nguyen (talk | contribs) |
||
| Line 86: | Line 86: | ||
<div class="accordionButton"><b>Output Amplification Model</b></div> | <div class="accordionButton"><b>Output Amplification Model</b></div> | ||
<div class="accordionContent"><br /><a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten"><b>Model based on Michaelis Menten Kinetics</b></a><br />Comparison between different amplification models (HIV1 and TEV) based on Michaelis Menten kinetics. However, Michaelis Menten kinetics does not apply to our system. Therefore, it had to be modelled from first principle (see below using law of mass action).<br /><img src="http://www.openwetware.org/images/0/03/Slide2.JPG" height=" | <div class="accordionContent"><br /><a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten"><b>Model based on Michaelis Menten Kinetics</b></a><br />Comparison between different amplification models (HIV1 and TEV) based on Michaelis Menten kinetics. However, Michaelis Menten kinetics does not apply to our system. Therefore, it had to be modelled from first principle (see below using law of mass action).<br /><a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten#HIV1"><img src="http://www.openwetware.org/images/0/03/Slide2.JPG" height="150" width="200" alt="Model using HIV1"/></a> <a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten#TEV"><img src="http://www.openwetware.org/images/4/48/TEV.jpg" height="150" width="200" alt="Model using TEV"/></a><br /><br /><a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action"><b>Model based on Law of Mass Action</b></a><br />Comparison between these 3 different models: Simple production, 1-step and 2-step amplification.<br /><a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action#Model_preA:_Simple_Production_of_Dioxygenase"><img src="http://www.openwetware.org/images/7/7f/Simple_production.JPG" height="100" width="300" alt="Simple Production"/></a> <a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action#Model_A:_Activation_of_Dioxygenase_by_TEV_enzyme"><img src="http://www.openwetware.org/images/1/1c/1-step_amplification.JPG" height="100" width="300" alt="1-step amplification"/></a> <a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action#Model_B:_Activation_of_Dioxygenase_by_TEV_or_activated_split_TEV_enzyme"><img src="http://www.openwetware.org/images/0/02/2-step_amplification.JPG" height="100" width="300" alt="2-step amplification"/></a><br /><a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Variables1"><b>Variables and Constants</b></a><br />Here are the variables and constants that are used in the Protein Display Model.<br /><br /></div> | ||
<div class="accordionButton"><b>Protein Display Model</b></div> | <div class="accordionButton"><b>Protein Display Model</b></div> | ||
Revision as of 16:14, 8 September 2010
These are our daily objectives.
Output Amplification Model
Comparison between different amplification models (HIV1 and TEV) based on Michaelis Menten kinetics. However, Michaelis Menten kinetics does not apply to our system. Therefore, it had to be modelled from first principle (see below using law of mass action).
 |
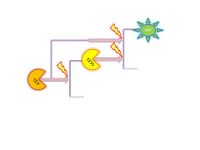 |
Comparison between these 3 different models (which are based on the law of mass action): Simple production, 1-step and 2-step amplification
 |
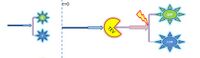 |
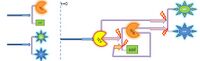 |
Here are the variables and constants that are used in the Output Amplification Model.
Protein Display Model
This is a model showing when the ComD receptor will be activated (after proteins have been cleaved).
Here are the variables and constants that are used in the Protein Display Model.
Feedback from Wetlab
<html> <head> <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.4.2/jquery.min.js"></"></script> <script type="text/javascript"> $(document).ready(function() {
$('div.accordionButton').click(function() {
$(this).next().slideToggle('normal'); });
$("div.accordionContent").hide();
});
</script> <style type="text/css">
- wrapper {
width: 1000px; margin-left: auto; margin-right: auto; }
.accordionButton { width: 1000px;
height: 20px;
float: left; text-align: center;
display: block;
background: #FFCC33;
border-bottom: 1px solid #FFFFFF; cursor: pointer;
padding: 10px;
}
.accordionContent { width: 1000px; float: left;
text-align: center;
background: #FFFF99; display: none;
padding:10px;
} </style> </head> <body>
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Objectives">Here are our daily objectives.</a>
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten">Model based on Michaelis Menten Kinetics</a>
Comparison between different amplification models (HIV1 and TEV) based on Michaelis Menten kinetics. However, Michaelis Menten kinetics does not apply to our system. Therefore, it had to be modelled from first principle (see below using law of mass action).
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten#HIV1"><img src="http://www.openwetware.org/images/0/03/Slide2.JPG" height="150" width="200" alt="Model using HIV1"/></a> <a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Michaelis_Menten#TEV"><img src="http://www.openwetware.org/images/4/48/TEV.jpg" height="150" width="200" alt="Model using TEV"/></a>
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action">Model based on Law of Mass Action</a>
Comparison between these 3 different models: Simple production, 1-step and 2-step amplification.
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action#Model_preA:_Simple_Production_of_Dioxygenase"><img src="http://www.openwetware.org/images/7/7f/Simple_production.JPG" height="100" width="300" alt="Simple Production"/></a> <a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action#Model_A:_Activation_of_Dioxygenase_by_TEV_enzyme"><img src="http://www.openwetware.org/images/1/1c/1-step_amplification.JPG" height="100" width="300" alt="1-step amplification"/></a> <a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Mass_Action#Model_B:_Activation_of_Dioxygenase_by_TEV_or_activated_split_TEV_enzyme"><img src="http://www.openwetware.org/images/0/02/2-step_amplification.JPG" height="100" width="300" alt="2-step amplification"/></a>
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Variables1">Variables and Constants</a>
Here are the variables and constants that are used in the Protein Display Model.
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Protein_Display">Protein Display Model</a>
This is a model showing when the ComD receptor will be activated (after proteins have been cleaved).
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Variables2">Variables and Constants</a>
Here are the variables and constants that are used in the Protein Display Model.
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Experiments1">Experiments for Output Amplification Model</a>
<a href="http://www.openwetware.org/wiki/IGEM:Imperial/2010/Modelling">Experiments for Protein Display Model</a>
</body> </html>