IGEM:IMPERIAL/2009/M3/Modelling/old/rubbish
Old stuff - to be tidied up
Then tidy it up!!! move this to another page please
1) Promoter characterisation
To determine how the PoPS Output of the promoters varies with time.
For constitutive promoter
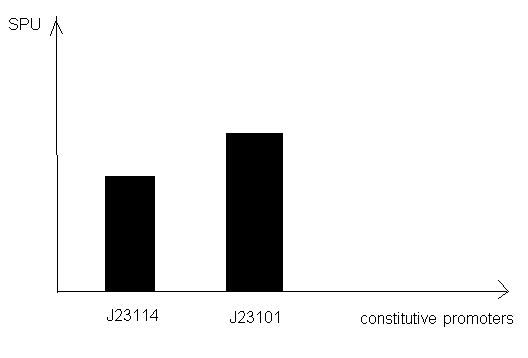
2) Thermoinduction
- Model how promoter activity is affected by temperature.
An example of a graph to be obtained.

what is mesured here????
This can be further correlated to explaining models on how cell death (cfu) is affected by temperature.
- Modelling cell growth with cI857 | Paper
3) Killing strategy
Cell staining (rate of killing)
From the plot of the percent live cells over time, we can determine the rate at which the cells are being killed. From here, we can model the action of the enzymes in terms of efficiency and time. We will hopefully be able to confidently predict cell death over a certain a time period.
Restriction Enzymes and Methylation
In our assays, we have determined quantitatively the enzymatic activity of our restriction enzymes and methylases. The balance between the amount of methylases needed to protect against the restriction enzymes, and the amount of methylase that will stifle restriction enzyme activity, can be modelled. From our in vitro derived models, we will hopefully come up with an optimum amount of methylation for our setup as well as after varying the promoter leakiness.

4)Characterization of enzymes in response to temperature
Based on the work from IGEM Delft 2008 Modelling, the team did some interesting characterization of enzymes in response to temperature.
They used hill type differential equations with temperature thresholds, and then fitted their results to work out the constants using data from the lab.
Similar work could be done in our project to characterize our restriction enzymes.
For more background, check wiki page and several links on optimization.
