BioSysBio:abstracts/2007/John Pinney
SHARKview: a tool for the visualization of systems biology data
Authors: John M Starkovich, Peter Li, David L Robertson and John W Pinney
Affiliations: University of Manchester
Contact: john.pinney@manchester.ac.uk
Keywords: 'Systems Biology' 'BioPAX' 'SBML' 'visualization'
Background/Introduction
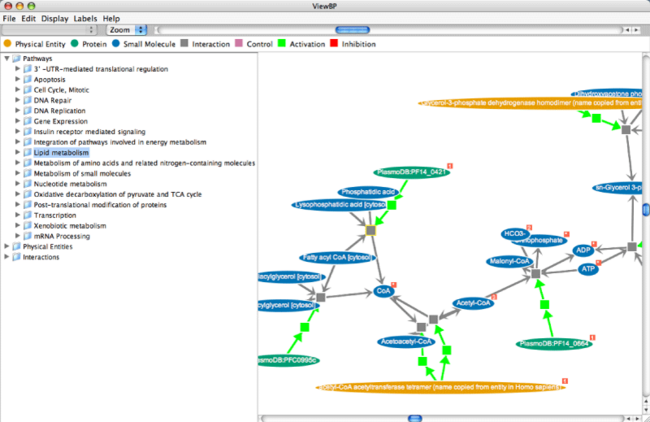
SHARKview is a freely available program to enable the interactive exploration of biological models encoded in the BioPAX or SBML exchange formats. This includes the ability to browse a model by pathway, physical entity, or interaction as well as the ability to overlay quantitative data such as gene expression levels onto the components of a model.
Results
Using Saccharomyces cerevisiae models obtained from online resources in conjunction with expression data from several different studies, we show how SHARKview can be used to visualise mRNA expression level changes for the proteins composing a pathway. Pathways analysed include both metabolic and genetic regulatory pathways.
Conclusion
Beyond simply displaying the model structure, the facility to incorporate and visualise transcriptomic, proteomic or metabolomic data in the context of a number of different cellular pathways will make SHARKview a very useful tool for systems biologists.
Images/Tables