BioMicroCenter:FAQ
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |
CONTACT
STOP BY: The BioMicro Center is located at 68-322 on the MIT campus.
EMAIL: The BioMicro staff can be emailed at biomicro@mit.edu
PHONE: The main lab number is 617-715-4533
Our director, Stuart Levine, can be reached at 617-452-2949.
DEFINITIONS
CORE LABS
The BioMicro Center is NOT an MIT institutional core, but a departmental resource. Departments that fund the operation of the BioMicro Center are given priority for all resources within the Center. Several labs that have provided major equipment donations to the BioMicro Center are also considered to be CORE. The BioMicro Center is funded by:
- The Department of Biology
- The Department of BioEngineering
- The David H. Koch Institute for Integrative Cancer Research
- MIT Center for Environmental Health Sciences
ASSISTED versus WALKUP SERVICES
Services available in the BioMicro Center are offered either as assisted or as walk-up. Walk-up Services are those where the Center provides training and maintenance of equipment and may also provide some consumables. Scheduling for Walk-up services is available through the calendar functions on ilabs and are almost exclusively limited to MIT users except in rare cases. Non-MIT users should contact biomicro@mit.edu to schedule walk-up equipment.
Assisted services are those where samples are delivered to the BioMicro Center and are analyzed by Center staff. These are set up using the FORMS on this site or the "Request Services" tab in ilabs. Assisted services cannot typically be "scheduled" with rare exceptions.
STANDARD versus HIGH THROUGHPUT LIBRARY PREPARATION
Standard library preparation is designed to maximize successful production of libraries and is used for routine work and should also be used for all precious samples. Standard library preparation includes initial sample quality control as well as at least one repeat for each library preparation if required due to sample failure. Standard library preparation is charged on a per sample basis. Standard libraries may be prepared by hand or using automation depending on throughput of the core.
High Throughput library (HTL) preparation is designed to minimize cost and has many differences from standard library preparation as a result. HTLs costs do NOT include initial sample quality control nor repeats of failures of individual samples. As such, we very strongly discourage HTL library prep for precious samples or experiments with limited replicates (N<=3). Replicate counts of 4 are considered the minimum with 6 being recommended where possible. Some methods, such as HT3DGE, can never repeat failed samples, while others can be reproduced by hand, though that does create a technical variable that can impact analysis. HTLs are almost always produced using liquid handlers, typically using miniaturization, and may require submission of the samples in specific formats (noted on each library prep type). HTLs are spot checked for quality after library production. Full quality control before and after experiments as well as sample arraying and sample cleaning is available at an additional charge.
We have published a manuscript highlighting some of the methods on JBT.
A webinar by our director for SPT Labtech can be found here
ILABS
The BioMicro Center uses ilabs for all internal submissions and for scheduling. All users must use ilabs IF:
- They are from MIT (including Whitehead Institute or Broad Institute with MIT appointments)
- They are using an MIT cost object to pay for the services
Users not from MIT should never use ilabs for submission to the BioMicro Center
Users from labs new to MIT may have a brief grace period to not use ilabs while their labs are set up
Users paying with POs may either use paper forms or ilabs but MUST bring the PO number with submission. We will use ilabs to import the project into our LIMS and close the project on ilabs without billing it.
APPROVAL
IMPORTANT: The BioMicro Center prioritizes timely delivery of genomic data. As such, WE DO NOT HOLD PROJECTS WAITING FOR FINANCIAL APPROVAL!. If a project is not rejected by the submitting lab in a timely manner, we will proceed with the project and recharge normally.
COST OBJECTS
The BioMicro Center does not control which cost objects are available to you for your project. If no cost object is available, you will NOT be able to submit via ilabs. Cost objects are assigned to you in ilabs by your PI or a designated administrator. Having permission to spend on a cost object through MIT is NOT automatically passed to ilabs and ilabs must be set up separately.
DOWNLOADING DATA
You will be notified by email that your data is ready. The data will be placed on one of our servers for you the download. All data is typically retained for 60 days. We STRONGLY encourage you to keep a local copy of your data as soon as possible. Contact bmc-data@mit.edu if you have any issues.
MIT / MIT-CERTIFICATE + VPN
| LURIA |
|
| Windows/Mac SFTP client |
|
| Web Download (MIT certificate required) |
https://bmc-data.mit.edu/[LAB]/[PROJECT] |
NON-MIT
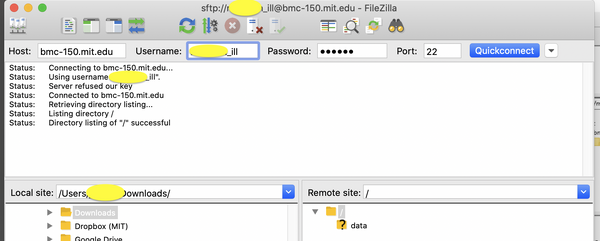
Non-MIT users should use a SFTP client to connect to bmc-150.mit.edu with credentials emailed to you. SSH or SCP is not supported.
| WINDOWS/MAC |
|
| UNIX / LINUX |
|
Please contact bmc-data@mit.edu if you have difficulty obtaining your data.
Read archival
Users are strongly encouraged to retain a local copy of their data as early as possible after sequencing result delivery.
After 60 days, fastq files will be compressed on the server. Bam files, containing reads aligned either to the genome mentioned in the sample submission form or phiX (a phage genome), will be deleted to save storage space.
Older files: Prior to 2020, bam files were maintained as archival files. Fastq files can be regenerated from the bam files (samtools sort -n to sort bam files by read names, and converted back to fastq files using bedtools bamToFastq).
UPLOADING DATA
To upload your data, you can access our file server named bmc-opendata.mit.edu via the sftp protocol.
- Request SFTP account with projects and lab information for storage setup
- Use a SFTP client (such as WinSCP, FileZilla, PuTTY and Cyberduck for Windows/Mac or sftp command for Linux)
NON MIT USERS
Do you take samples from outside MIT?
The BioMicro Center is built to serve the MIT community. As such, members of the MIT community have priority on all of our services. However, if we have extra capacity, we are happy to make it available to scientists not affiliated with MIT, with the caveats that MIT samples *always* have priority and that access is finite. The BioMicro Center retains the right to refuse any sample.
How can we ship samples to you?
Please email biomicro@mit.edu to arrange a drop off date and time. DNA samples should be shipped at 4C and RNA samples should be shipped on dry ice.Samples should be submitted with a completed order form and shipped by overnight delivery to:
MIT BioMicro Center 31 Ames Street, Building 68-322 Cambridge, MA 02139
An electronic copy of the full PO should be emailed to biomicro@mit.edu
The pricing form says "NA". What does that mean?
Some of our services are specifically restricted to the MIT community and we cannot offer them to outside users. Others are restricted to academic labs. Please email us at biomicro@mit.edu if you have any questions.
What is your billing address?
Payments should be made to our finance office at:
Massachusetts Institute of Technology Biology Finance Office, Attn: Alison Salie 77 Massachusetts Avenue, 68-157 Cambridge, MA 02139
Do you accept credit cards?
We are unable to accept payment by credit card.
MIT W9