Wwwblast: Difference between revisions
| (11 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
* It is more difficult to use when dealing with large numbers of sequences, and is not amenable to parsing using a [[BioPerl]] parser, for instance. | * It is more difficult to use when dealing with large numbers of sequences, and is not amenable to parsing using a [[BioPerl]] parser, for instance. | ||
[[Image:Wwwblast.png|thumb|right|Webpage used to submit BLAST queries with wwwblast.]] | [[Image:Wwwblast.png|thumb|right|Webpage used to submit BLAST queries with wwwblast.]] | ||
It should perhaps be noted that the author of this tutorial now no longer uses wwwblast, but [http://www.sequenceserver.com SequenceServer] instead, since it is much easier to setup and use (in the author's opinion). | |||
=Setting up a wwwblast server with the Apache web server= | =Setting up a wwwblast server with the Apache web server= | ||
| Line 65: | Line 67: | ||
A binary BLAST database is a collection of multiple files (.nhr, .nin and .nsq files for nucleotide databases). They must be created from a fasta file in a terminal, using the BLAST+ toolkit, available from [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download NCBI] ([http://www.ncbi.nlm.nih.gov/pubmed/20003500 PubMed citation]). The legacy BLAST toolkit can be used to achieve the same goal, though the command line syntax differs. | A binary BLAST database is a collection of multiple files (.nhr, .nin and .nsq files for nucleotide databases). They must be created from a fasta file in a terminal, using the BLAST+ toolkit, available from [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download NCBI] ([http://www.ncbi.nlm.nih.gov/pubmed/20003500 PubMed citation]). The legacy BLAST toolkit can be used to achieve the same goal, though the command line syntax differs. | ||
NOTE: If you are planning on using [[Blast_link]], follow the instructions on that | NOTE: If you are planning on using [[Blast_link]], follow the instructions on that wiki page for this section, as it differs slightly. | ||
After copying the fasta file (called for example 'my_nucleotide_sequences.fasta') to the db directory of the wwwblast installation (e.g. /Users/ben/Sites/blast/db), enter the following in a terminal: | After copying the fasta file (called for example 'my_nucleotide_sequences.fasta') to the db directory of the wwwblast installation (e.g. /Users/ben/Sites/blast/db), enter the following in a terminal: | ||
| Line 116: | Line 118: | ||
=Troubleshooting= | =Troubleshooting= | ||
==Cannot find makeblastdb== | |||
Specify the full path to the makeblastdb executable. TODO. | |||
==No hits found== | |||
[[Image:Bad_no_hits.png|thumb|right|A false "no hits found", where there actually the binary blast database is not in the correct place.]] | [[Image:Bad_no_hits.png|thumb|right|A false "no hits found", where there actually the binary blast database is not in the correct place.]] | ||
[[Image:Good_no_hits.png|thumb|right|A true "no hits found", where the binary database is correctly configured.]] | [[Image:Good_no_hits.png|thumb|right|A true "no hits found", where the binary database is correctly configured.]] | ||
Somewhat misleadingly, if the binary databases are not in the correct place, but blast.rc is specified correctly, then wwwblast will not warn of this directly. Instead, it will display "***** no hits found *****" but not complete the web page by adding database statistics. If there really are no hits found, the statistics about the database will be displayed below. See the two contrasting pictures to the right of this text. | Somewhat misleadingly, if the binary databases are not in the correct place, but blast.rc is specified correctly, then wwwblast will not warn of this directly. Instead, it will display "***** no hits found *****" but not complete the web page by adding database statistics. If there really are no hits found, the statistics about the database will be displayed below. See the two contrasting pictures to the right of this text. | ||
==Internal Server Error== | |||
If a webpage is encountered that says "Internal Server Error", something has gone wrong while the webserver was running wwwblast. To find out more about this error, look at the end of the apache error log. The error log is a file, which might be /var/log/httpd/error_log or /var/log/apache2/error.log. | |||
==Exec of blast.cgi failed== | |||
If you end up with an error like this: | |||
<pre> | |||
No such file or directory: exec of ‘/var/www/blast/blast.cgi’ failed, referer: http://localhost/blast/blast.html | |||
</pre> | |||
This might be because: | |||
# csh is not installed. This is unlikely to be the case on OSX because csh comes installed by default | |||
# blast.cgi does not have execute permissions. | |||
=Missing Features of WWW-BLAST= | =Missing Features of WWW-BLAST= | ||
| Line 126: | Line 143: | ||
* '''No security'''. By the methods shown above, the sequences in the database are accessible to everyone on the internet. A username and password system may be added by configuring [[apache basic authentication]]. | * '''No security'''. By the methods shown above, the sequences in the database are accessible to everyone on the internet. A username and password system may be added by configuring [[apache basic authentication]]. | ||
* '''No dead-easy installer'''. If you've followed the above tutorial, you'll realise that installation of www-blast is more complicated than installing the average program. Sorry about this one folks, but there's no known fix. | * '''No dead-easy installer'''. If you've followed the above tutorial, you'll realise that installation of www-blast is more complicated than installing the average program. Sorry about this one folks, but there's no known fix. | ||
* '''Cannot search multiple databases simultaneously'''. The database selection is a drop-down box, not a list of checkboxes. So only one database can be searched at a time. | |||
=See also= | |||
* [[BLAST]] - the underlying algorithm that wwwblast uses | |||
* [[Blast_link]] - an extension of wwwblast that allows access to the full sequences of the hits | |||
* [[apache basic authentication]] - a way to password-protect the wwwblast webpage | |||
=Alternatives= | |||
* [http://www.sequenceserver.com/ SequenceServer] An alternative to wwwblast that is much easier to set up, use, and is actively maintained software. It is free for academic/non-profit/educational institutions. | |||
=Acknowledgements= | =Acknowledgements= | ||
Creation of this wiki page was funded by the [http://www.mol-palaeo.de Molecular Geo- and Palaeobiology Lab] of the Ludwig-Maximilians-Universität (LMU). | Creation of this wiki page by [[User:Ben_Woodcroft]] was funded by the [http://www.mol-palaeo.de Molecular Geo- and Palaeobiology Lab] of the Ludwig-Maximilians-Universität (LMU). | ||
[[Category:Protocol]] | |||
[[Category:In silico]] | |||
[[Category:DNA]] | |||
[[Category:Protein]] | |||
[[Category:Sequence analysis]] | |||
Latest revision as of 17:23, 3 January 2012
A wwwblast installation allows a researcher to use BLAST to search a sequence database using a graphical user interface. There are several differences when compared to using BLAST from the command line (as is described in Wikiomics:BLAST_tutorial for instance):
- After setup, researchers use the program by accessing a web page, not using the command line. This can make it more suitable for computer-shy biologists.
- The output includes a diagrammatic overview of the BLAST hits' coverage of the query sequence, whereas command line BLAST does not generate this.
- It is more difficult to use when dealing with large numbers of sequences, and is not amenable to parsing using a BioPerl parser, for instance.

It should perhaps be noted that the author of this tutorial now no longer uses wwwblast, but SequenceServer instead, since it is much easier to setup and use (in the author's opinion).
Setting up a wwwblast server with the Apache web server
Install the pre-requisites
wwwblast requires csh and apache to be installed. On OSX, these are installed by default. It can be setup on windows computers, but that is not dealt with here.
Download and extract
Download the wwwblast program from the NCBI FTP site.
Extract the contents of the downloaded archive into the default apache directory (the DocumentRoot, in apache parlance):
- On OSX, this site is /Users/ben/Sites (replacing ben with your login name)
- On Ubuntu and other linux distributions, use /var/www
You can extract using a graphical user interface, but also on the command line. For instance, on Linux,
$ cd /var/www $ sudo tar xzf /home/ben/Downloads/wwwblast-2.2.24-ia32-linux.tar.gz
Afterwards, the folder /Users/<username>/Sites/blast should exist (or the equivalent on other operating systems). Technically there is no reason it cannot be in any directory in the DocumentRoot e.g. /Users/ben/Sites/myblasts/blast, but the simplest directory is assumed in this howto. For the rest of the tutorial the directory /User/ben/Sites/blast is assumed to be base directory of the blast installation, and 'ben' is assumed to be the user logged into the computer.
Turning on the webserver
Enabling the Apache web server is different on different platforms. On OSX, enable the personal web sites option (System Preferences - Network/Sharing - Personal Web Sharing). On Ubuntu, install the apache2 package.
When this step is correctly carried out, the webpage http://localhost should work.
Modifying the apache configuration file
Add the following text entry to the apache config file. You will need administrator privileges.
- On OSX (only older versions?) this file is /etc/httpd/users/ben.conf
- On Ubuntu and some other Linux distributions, create a new file /etc/apache2/conf.d/blast.conf
# Added below to get wwwblast to work AddHandler cgi-script .cgi <Directory "/Users/ben/Sites/blast"> Options FollowSymLinks +ExecCGI +Indexes </Directory>
After saving the file, restart the apache webserver. The simplest way is to restart the computer. If restarting the computer is not possible you can restart the server using apache2ctl, with administrator privileges:
$ sudo apache2ctl graceful
After restarting, you should be able to run a blast against the default test databases.
- On OSX, the URL will be http://localhost/~ben/blast/blast.html.
- On Linux, the URL will be http://localhost/blast/blast.html.
You can use the sequence TACTGTTATCGATCCGGTCGAAAAACTGCTGGCAGTGGGGCATTACCTCGAATCTACCGTCGATATTGCT to test, against test_na_db using blastn. You should get a hit, not a half-blank page.
Getting the BLAST overview image to work
If there is no image showing how the blast sequence worked, and instead in its place was the an error message similar to below though with the numbers being different:
fail to open file TmpGifs/1804289383790.gif
This can be fixed by giving every user on the computer write access to the TmpGifs folder. In a terminal (on Linux, need administrator privileges):
$ chmod o+w /Users/ben/Sites/blast/TmpGifs/
There should be no output from this command. If the permissions have been changed correctly, the overview image should work.
Creating custom databases
By default, wwwblast only comes with a few databases that are generally not useful. Instead the researcher wants to BLAST against sequences generated in their own lab, for instance. To make a custom sequence database from a FASTA file, 3 steps are required: converting the fasta file into a binary blast database, changing the blast.html so that the database can be selected from the drop-down menu, and modifying blast.rc.
Converting a fasta file into a binary BLAST database
A binary BLAST database is a collection of multiple files (.nhr, .nin and .nsq files for nucleotide databases). They must be created from a fasta file in a terminal, using the BLAST+ toolkit, available from NCBI (PubMed citation). The legacy BLAST toolkit can be used to achieve the same goal, though the command line syntax differs.
NOTE: If you are planning on using Blast_link, follow the instructions on that wiki page for this section, as it differs slightly.
After copying the fasta file (called for example 'my_nucleotide_sequences.fasta') to the db directory of the wwwblast installation (e.g. /Users/ben/Sites/blast/db), enter the following in a terminal:
$ cd /Users/ben/Sites/blast/db $ makeblastdb -in my_nucleotide_sequences.fasta -dbtype nucl Building a new DB, current time: 09/23/2010 14:12:18 New DB name: my_nucleotide_sequences.fasta New DB title: my_nucleotide_sequences.fasta Sequence type: Nucleotide Keep Linkouts: T Keep MBits: T Maximum file size: 1073741824B Adding sequences from FASTA; added 1620 sequences in 0.207906 seconds.
For amino acid sequence fasta files, use '-dbtype prot' instead of '-dbtype nucl'
Adding the custom database to the drop-down box
To search against a custom database, it must be able to be selected from the drop-down box in blast.html. To add a database to the drop-down box, modify blast.html in a text editor, such as TextWrangler on OSX, notepad on windows, or gedit on linux. An example:
<select name = "DATALIB">
<option VALUE = "my_nucleotide_sequences.fasta"> My nucleotide sequences
<option VALUE = "test_na_db"> test_na_db
<option VALUE = "test_aa_db"> test_aa_db
</select>
After the HTML has been modified correctly, the custom database should be able to be found when viewing the blast page with an internet browser, for example at http://localhost/blast/blast.html.
Modifying blast.rc
Without modifying the blast.rc file correctly for a given database, the following error will appear when a blast is run:
Error 9 in submitting BLAST query Short error description: The combination of database and program, that you provided in your message is invalid or not acceptable by BLAST search system. Please look at current possible combinations in BLAST help.
To fix it, change the blast.rc file, which is in the blast folder to something akin to below:
blastn test_na_db my_nucleotide_sequences.fasta blastp test_aa_db blastx test_aa_db tblastn test_na_db my_nucleotide_sequences.fasta tblastx test_na_db my_nucleotide_sequences.fasta
As many databases can be added space separated as required after each program.
After completing this step, you should be able to run your blast from http://localhost/blast/blast.html. It is a good idea to run a positive control. Take a section of sequence from the fasta file of the database you are blasting against, and query the database with it. A positive control is a good idea because wwwblast reporting "no hits found" can be misleading - see the troublshooting section.
Troubleshooting
Cannot find makeblastdb
Specify the full path to the makeblastdb executable. TODO.
No hits found

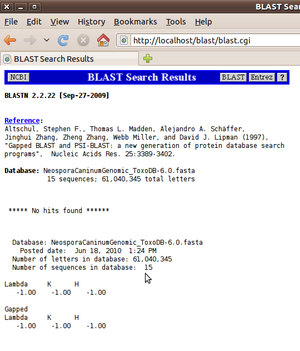
Somewhat misleadingly, if the binary databases are not in the correct place, but blast.rc is specified correctly, then wwwblast will not warn of this directly. Instead, it will display "***** no hits found *****" but not complete the web page by adding database statistics. If there really are no hits found, the statistics about the database will be displayed below. See the two contrasting pictures to the right of this text.
Internal Server Error
If a webpage is encountered that says "Internal Server Error", something has gone wrong while the webserver was running wwwblast. To find out more about this error, look at the end of the apache error log. The error log is a file, which might be /var/log/httpd/error_log or /var/log/apache2/error.log.
Exec of blast.cgi failed
If you end up with an error like this:
No such file or directory: exec of ‘/var/www/blast/blast.cgi’ failed, referer: http://localhost/blast/blast.html
This might be because:
- csh is not installed. This is unlikely to be the case on OSX because csh comes installed by default
- blast.cgi does not have execute permissions.
Missing Features of WWW-BLAST
Even when properly setup, the default installation of wwwblast has some missing features;
- No access to the entire hit sequence. When the query sequence has a hit against the database, there is no way to retrieve the entire hit sequence for further analysis. This may be fixed by installing the blast_link add-on.
- No security. By the methods shown above, the sequences in the database are accessible to everyone on the internet. A username and password system may be added by configuring apache basic authentication.
- No dead-easy installer. If you've followed the above tutorial, you'll realise that installation of www-blast is more complicated than installing the average program. Sorry about this one folks, but there's no known fix.
- Cannot search multiple databases simultaneously. The database selection is a drop-down box, not a list of checkboxes. So only one database can be searched at a time.
See also
- BLAST - the underlying algorithm that wwwblast uses
- Blast_link - an extension of wwwblast that allows access to the full sequences of the hits
- apache basic authentication - a way to password-protect the wwwblast webpage
Alternatives
- SequenceServer An alternative to wwwblast that is much easier to set up, use, and is actively maintained software. It is free for academic/non-profit/educational institutions.
Acknowledgements
Creation of this wiki page by User:Ben_Woodcroft was funded by the Molecular Geo- and Palaeobiology Lab of the Ludwig-Maximilians-Universität (LMU).