Work Area: Difference between revisions
Superphage (talk | contribs) |
Superphage (talk | contribs) |
||
| Line 282: | Line 282: | ||
[[Image: | [[Image:Enzymes5.jpg]] | ||
Table X. | Table X. | ||
Revision as of 12:20, 15 August 2007
Papers to cite
biosensors
- Daunert S, Barrett G, Feliciano JS, Shetty RS, Shrestha S, and Smith-Spencer W. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem Rev. 2000 Jul 12;100(7):2705-38. DOI:10.1021/cr990115p |
- Yagi K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl Microbiol Biotechnol. 2007 Jan;73(6):1251-8. DOI:10.1007/s00253-006-0718-6 |
- Harms H, Wells MC, and van der Meer JR. Whole-cell living biosensors--are they ready for environmental application?. Appl Microbiol Biotechnol. 2006 Apr;70(3):273-80. DOI:10.1007/s00253-006-0319-4 |
- Gu MB, Min J, and Kim EJ. Toxicity monitoring and classification of endocrine disrupting chemicals (EDCs) using recombinant bioluminescent bacteria. Chemosphere. 2002 Jan;46(2):289-94. DOI:10.1016/s0045-6535(01)00081-9 |
Papers cited by Forrest's writings
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, and O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003 Oct 16;425(6959):686-91. DOI:10.1038/nature02026 |
- Pajot-Augy E, Crowe M, Levasseur G, Salesse R, and Connerton I. Engineered yeasts as reporter systems for odorant detection. J Recept Signal Transduct Res. 2003;23(2-3):155-71. DOI:10.1081/rrs-120025196 |
Biofuel
The need for alternative fuels has increased considerably in the recent years due to rising fossil fuel prices and worldwide attention towards environmental sustainability. While battery technology has steadily progressed, electrochemical energy storage still cannot (and may never) compete with liquid or gaseous fuels on energy density.
Among the promising candidates for alternative fuels are biofuels and hydrogen. Biofuels -- fuels produced from crop-based carbohydrates -- include bioethanol from fermentable corn or switchgrass sugars and biodiesel from plant oils (torney). Numerous groups have employed recombinant and metabolic engineering to develop microorganisms that produce biofuels. For example, a recombinant Saccharomyces yeast strain has been engineered to break down cellulose and ferment ethanol from xylose, a pentose commonly found in renewable lignocellulosic biomass such as waste paper. This was achieved by introducing genes from P. stipitis and S. cerevisiae for their xylose-fermenting ability, and displaying on the cell surface a cellooligosaccharide-degrading fusion protein from an A. aculeatus gene and α-agglutinin (Katahira). Commercial entities pursuing biofuel applications of synthetic biology include California startups Amyris Biotechnologies, LS9, and Synthetic Genomics.
- Torney F, Moeller L, Scarpa A, and Wang K. Genetic engineering approaches to improve bioethanol production from maize. Curr Opin Biotechnol. 2007 Jun;18(3):193-9. DOI:10.1016/j.copbio.2007.03.006 |
- Prasad D, Arun S, Murugesan M, Padmanaban S, Satyanarayanan RS, Berchmans S, and Yegnaraman V. Direct electron transfer with yeast cells and construction of a mediatorless microbial fuel cell. Biosens Bioelectron. 2007 May 15;22(11):2604-10. DOI:10.1016/j.bios.2006.10.028 |
- Okuda N, Ninomiya K, Takao M, Katakura Y, and Shioya S. Microaeration enhances productivity of bioethanol from hydrolysate of waste house wood using ethanologenic Escherichia coli KO11. J Biosci Bioeng. 2007 Apr;103(4):350-7. DOI:10.1263/jbb.103.350 |
- Henstra AM, Sipma J, Rinzema A, and Stams AJ. Microbiology of synthesis gas fermentation for biofuel production. Curr Opin Biotechnol. 2007 Jun;18(3):200-6. DOI:10.1016/j.copbio.2007.03.008 |
- Katahira S, Mizuike A, Fukuda H, and Kondo A. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol. 2006 Oct;72(6):1136-43. DOI:10.1007/s00253-006-0402-x |
- Bro C, Regenberg B, Förster J, and Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab Eng. 2006 Mar;8(2):102-11. DOI:10.1016/j.ymben.2005.09.007 |
- Hill J, Nelson E, Tilman D, Polasky S, and Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci U S A. 2006 Jul 25;103(30):11206-10. DOI:10.1073/pnas.0604600103 |
- Zhang YH, Evans BR, Mielenz JR, Hopkins RC, and Adams MW. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS One. 2007 May 23;2(5):e456. DOI:10.1371/journal.pone.0000456 |
-
=
-
=
-
=
Papers baby wants
- Lu TK and Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11197-202. DOI:10.1073/pnas.0704624104 |
Used in paper already
- Keane A, Phoenix P, Ghoshal S, and Lau PC. Exposing culprit organic pollutants: a review. J Microbiol Methods. 2002 Apr;49(2):103-19. DOI:10.1016/s0167-7012(01)00382-7 |
Bacterial plastics
- Ward PG, Goff M, Donner M, Kaminsky W, and O'Connor KE. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ Sci Technol. 2006 Apr 1;40(7):2433-7. DOI:10.1021/es0517668 |
- Liu SJ and Steinbüchel A. A novel genetically engineered pathway for synthesis of poly(hydroxyalkanoic acids) in Escherichia coli. Appl Environ Microbiol. 2000 Feb;66(2):739-43. DOI:10.1128/AEM.66.2.739-743.2000 |
Chronology of biosensors
1990
1991
1992
1993
1994
Pentachlorophenol
- Van Dyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, and LaRossa RA. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994 May;60(5):1414-20. DOI:10.1128/aem.60.5.1414-1420.1994 |
1995
1996
1997
1998
1999
2000
2001
2002
Endocrine disrupting chemicals (EDCs)
- Gu MB, Min J, and Kim EJ. Toxicity monitoring and classification of endocrine disrupting chemicals (EDCs) using recombinant bioluminescent bacteria. Chemosphere. 2002 Jan;46(2):289-94. DOI:10.1016/s0045-6535(01)00081-9 |
Herbicides
- Shao CY, Howe CJ, Porter AJ, and Glover LA. Novel cyanobacterial biosensor for detection of herbicides. Appl Environ Microbiol. 2002 Oct;68(10):5026-33. DOI:10.1128/AEM.68.10.5026-5033.2002 |
Genotoxic agents -- MMC, MNNG, NA
- Weisweiler P and Schwandt P. Biological action of the lipotrophic peptides A and B from pig pituitary glands. Acta Endocrinol (Copenh). 1975 May;79(1):34-42. DOI:10.1530/acta.0.0790034 |
2003
2004
2005
Chlorinated aliphatic hydrocarbons (CAHs) -- trichloroethene (TCE) in ground water
- Bhattacharyya J, Read D, Amos S, Dooley S, Killham K, and Paton GI. Biosensor-based diagnostics of contaminated groundwater: assessment and remediation strategy. Environ Pollut. 2005 Apr;134(3):485-92. DOI:10.1016/j.envpol.2004.09.002 |
2006
Heavy metals
- Shao CY, Howe CJ, Porter AJ, and Glover LA. Novel cyanobacterial biosensor for detection of herbicides. Appl Environ Microbiol. 2002 Oct;68(10):5026-33. DOI:10.1128/AEM.68.10.5026-5033.2002 |
2007
Figures
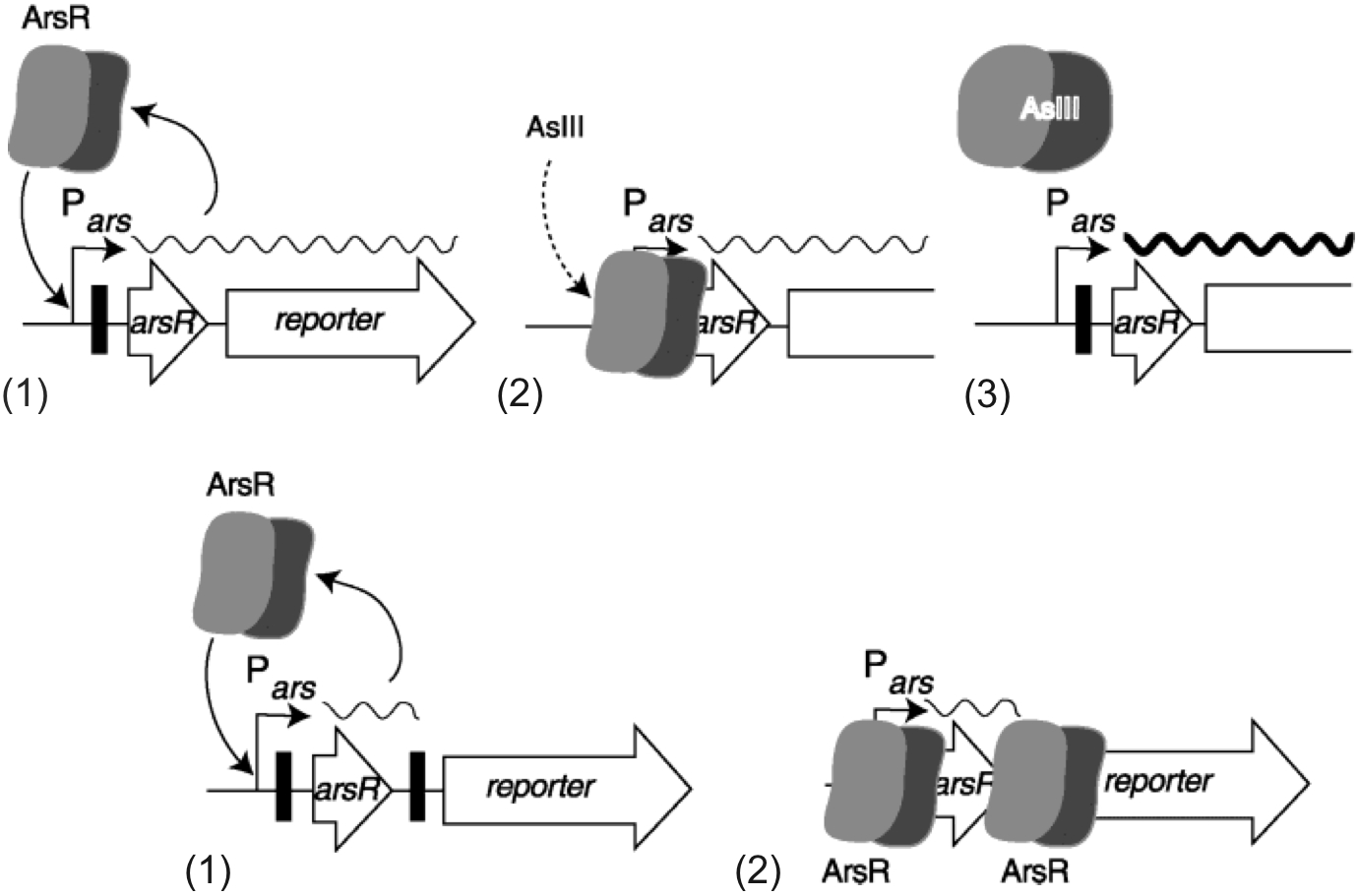
Figure 5. Schematic of arsenic biosensors. Whole-cell living bacterial biosensors for arsenite usually contain the arsR responsive promoter and arsR gene fused to a reporter gene; ArsR controls reporter gene expression. A: (1) With one ArsR binding site (black vertical bar), some background expression occurs from the arsR promoter (Pars) to synthesize ArsR itself (symbolized as a dimer) and consequently some reporter protein. (2) In the absence of arsenite (AsIII), ArsR binds to the promoter region. (3) But in the presence of arsenite, ArsR loses its affinity for its binding site and transcription from the arsR promoter will be very high (thick wave line). B: (1) With a secondary ArsR binding site downstream of arsR, the reporter protein is less frequently expressed as background (dotted wave line). (2) ArsR will bind to both DNA binding sites and lower the probability that RNA polymerase reads the reporter gene. In the presence of arsenite, ArsR will again lose its affinity, and expression from the reporter gene will be initiated. (From Stocker, 2003).
ADD TO TEXT: This dual binding site design works as two filter in series: If, for example, one filter (ArsR binding site) allows background expression 10% of the time, two such filters placed in series would allow expression only 1% of the time.
Example of Abstraction Hierarchy. The oscillator system is composed of three inverter devices, each of which is composed of a ribosome binding site part, a protein coding part, terminator parts, and a regulatory part. DNA-level researchers need only be concerned with creating a specific DNA sequence; part-level researchers need only be concerned with designing DNA sequences with a certain functionality; device-level researchers need only be concerned with combining existing parts to achieve a certain input-output response; and system-level researchers need only be concerned with combining existing devices to build a system with certain behaviors.
- Sørensen SJ, Burmølle M, and Hansen LH. Making bio-sense of toxicity: new developments in whole-cell biosensors. Curr Opin Biotechnol. 2006 Feb;17(1):11-6. DOI:10.1016/j.copbio.2005.12.007 |
Text
BS about education
The arsenic biosensor project serves as an inspiring preview of the future of environmental synthetic biology. As the Registry grows and commercial DNA synthesis becomes more affordable, we can expect even more impressive results from diverse teams of researchers across the world. Already, the openness and spirit of innovation of iGEM is motivating a new generation of scientists, engineers, business leaders, and legislators to look towards implementing synthetic biology to address global sustainability issues. With increasingly sophisticated synthetic biological systems and pathways for environmental sensing and remediation, fuel production, and biodegradable materials fabrication, [INSERT A CATCHY PHRASE such as "the future of the world is bright"]
Bioremediation
While it is valuable to detect environmental pollutants, removing them is just as important. Bioremediation--the process of utilizing organisms to rid an area of contaminants--has been improved throughout this decade by clever genetic engineering. For example, to enhance the bioaccumulation (i.e. absorption) of mercury by E. coli in soil and water, a de novo designed metal-binding peptide, (Glu-Cys)20Gly, has been expressed on the E. coli cell surface to enhance its mercury uptake (Bae et al., 2001). Further abstraction, decoupling, and standardization of bioengineering will likely accelerate future bacterial bioremediation research.
In another creative example, a biofilm consortium comprising engineered E. coli and P. putida was assembled to employ the pair's complementary metabolic pathways to biodegrade the organophosphate insecticide parathion (Gilbert et al., 2003). As synthetic biologists create more tools for developing cell-cell communication networks, environmental biotechnologists may soon be engineering remarkable bacterial consortia that respond to external conditions and work together to cleanse the environment with unprecedented effectiveness.
- Gilbert ES, Walker AW, and Keasling JD. A constructed microbial consortium for biodegradation of the organophosphorus insecticide parathion. Appl Microbiol Biotechnol. 2003 Mar;61(1):77-81. DOI:10.1007/s00253-002-1203-5 |
- Bae W, Mehra RK, Mulchandani A, and Chen W. Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl Environ Microbiol. 2001 Nov;67(11):5335-8. DOI:10.1128/AEM.67.11.5335-5338.2001 |
Biodegradable Plastics
Since synthetic polymers were first commercialized in the 1920s, they have found application in nearly every aspect of modern day life, including disposable bags, hard containers, electronics, wrappings, foam, laminated paper, fabrics, and many other ubiquitous items. In 1991 alone, fifteen billion pounds of polyethylene were consumed in the United States (chum91).
While cheap and versatile, synthetic polymers are becoming a significant environmental hazard because they can take many (up to several hundred) years to degrade naturally in landfill. Furthermore, synthetic polymers are typically derived from petroleum, a quickly diminishing resource that incurs significant environmental costs during extraction. For these reasons, there is a real need to produce biodegradable polymers from renewable resources.
Among several potentially viable biodegradable polymers, polyhydroxyalkanoates (PHAs) are especially promising because they can be produced in vivo by several species of microorganisms. Since polymers are commodity materials (i.e. highly cost competitive), biodegradable polymers must be priced comparably with synthetic polymers in order to survive on the marketplace. Synthetic biology can help by increasing the PHA yield and facilitating easier PHA extraction from the microorganisms.
Several species of bacteria––e.g. some in the archaebacterial family Halobacteriaceae (Hezayen02)--accumulate PHA granules when they experience a stress condition involving a nutrient imbalance such as excess carbon (Füchtenbusch98). However, since natural producers of PHAs tend to grow slowly, introducing PHA accumulation capabilities (i.e. genes encoding PHA synthase and other necessary enzymes) into a fast growing organism such as E. coli raises the overall yield rate and thereby lowers the cost of the polymer. And while engineered plant hosts may be more cost-effective for large scale PHA production because they collect energy from the sun and carbon from air, microbial systems offer more options for polymer composition and carbon source (e.g. polystyrene [ward06]) (aldor03).
PHA synthesis in microbes involves only a few enzymes (Fig. X [aldor03]), and genes encoding these enzymes can be introduced to recombinant bacteria from a number of organisms (Fig. X2 [suri07]). In the simplest pathway, the phaA gene encodes the enzyme, beta-ketothiolase, for the condensation of a pair of acetyl-CoA molecules to form acetoacetyl-CoA, which is then reduced to (R)-3-hydroxyacyl-CoA by the NADPH-cofactor-dependent acetoacetyl-CoA reductase encoded by the phaB gene. Lastly, the phaC gene encoded PHA synthase catalyzes the polymerization of (R)-3-hydroxyacyl-CoA monomers to PHA (aldor03).
Other pathways to supply (R)-3-hydroxyacyl-CoA monomers for PHA synthesis include fatty acid beta-oxidation, as found in bacteria such as P. oleovorans and P. fragii; and fatty acid de novo synthesis, as found in bacteria such as P. aeruginosa and P. putida. Both pathways have been successfully implemented in recombinant E. coli to allow for high rate production of various PHAs and PHA copolymers (suri07).
Mass-produced bacterial and plant-derived PHAs such as Mirel(TM) by Metabolix are just beginning to find commercial application. As synthetic biology matures and becomes more widely used in the engineering of biopolymers, we may see significant cost reductions and greater use of biodegradable polymers. In addition, advances in synthetic biology may also lead to the creation of useful metabolic and enzymatic pathways for the degradation and recycling of polymer wastes.
Figure X. Microbial metabolic pathways for PHA synthesis. Names of genes encoding key enzymes are boxed. Enzymes encoded by shown genes: PhaA, 3-ketothiolase; PhaB, (R)-3-ketoacyl-CoA reductase; PhaC, PHA synthase or polymerase; PhaG, (R)-3-hydroxyacyl ACP:CoA transacylase; PhaJ, (R)-specific enoyl-CoA hydratase. Dotted lines represent reactions in which intermediate metabolic steps are not shown.
Figure X2. pha gene operons in different microorganisms. (a) Complete phaCAB operon of R. eutropha; (b) interrupted locus of Z. ramigera; (c) locus with two polymerase subunits, phaC and phaE, of C. vinosum; (d) phaCJ operon of A. caviae for PHA copolymer formation; (e) phaC1ZC2D operon for PHA formation in P. oleovorans with two phaC genes; (f) pha locus with depolymerase (phaZ) between two polymerase subunits in P. aureofaciens and phaF gene situated downstream of phaC1ZC2D operon.
-
=
-
=
- Aldor IS and Keasling JD. Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol. 2003 Oct;14(5):475-83. DOI:10.1016/j.copbio.2003.09.002 |
- Suriyamongkol P, Weselake R, Narine S, Moloney M, and Shah S. Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants - a review. Biotechnol Adv. 2007 Mar-Apr;25(2):148-75. DOI:10.1016/j.biotechadv.2006.11.007 |
- Ward PG, Goff M, Donner M, Kaminsky W, and O'Connor KE. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ Sci Technol. 2006 Apr 1;40(7):2433-7. DOI:10.1021/es0517668 |
- Hezayen FF, Steinbüchel A, and Rehm BH. Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56. Arch Biochem Biophys. 2002 Jul 15;403(2):284-91. DOI:10.1016/s0003-9861(02)00234-5 |
- Steinbüchel A and Füchtenbusch B. Bacterial and other biological systems for polyester production. Trends Biotechnol. 1998 Oct;16(10):419-27. DOI:10.1016/s0167-7799(98)01194-9 |
Polymers from Biobased Materials By Helena L. Chum Published 1991 William Andrew Inc. Technology & Industrial Arts 169 pages ISBN 0815512716
Educational Initiatives
A significant result of synthetic biology is the democratization of bioengineering. Efforts in improving the abstraction, decoupling, and standardization of bioengineering have enabled novice researchers (even high school students) to create and utilize biological parts, devices, and systems.
In 2003, students at MIT participated in a month-long bioengineering mini course and created oscillator systems coupled to fluorescent reporters. The surprising success of this program led to the NSF-sponsored Synthetic Biology Competition (SBC) in 2004, in which five teams from within the United States designed and built genetically encoded machines using standard, interchangeable biological parts. The flagship product of this competition was UT Austin's biological photographic film, a biofilm with bacteria containing a light-detection device that produces PoPS in the presence of photons, as well as a reporting device that produces color in response to the PoPS. The light-detection device was created by combining a new light-detecting part (a bioengineered two-component signaling protein based on cph1 from Synechocystis) with a pre-existing BioBrick part; and the reporting device was created by combining three existing BioBricks parts.
To encourage further student participation in synthetic biology, researchers Drew Endy, Thomas Knight, and Randy Rettberg founded the international Genetically Engineered Machine (iGEM) competition in 2005, which in its first year involved thirteen teams competing from four countries. Through media publicity and rigorous outreach efforts by the competition's staff, iGEM 2006 had 37 teams from fifteen countries develop biological systems and present their results at a judged "Jamboree" event at MIT. To this date, 57 teams have registered to compete in iGEM 2007. Furthermore, several universities have initiated synthetic biology related courses, labs, and iGEM-style mini competitions.
Among the 37 projects in iGEM 2006, four were environmental applications of synthetic biology. Prairie View A&M University took steps towards a metal contamination detection; Mississipi State Univerity designed a hydrogen detecting part for future use in hydrogen-producing bacteria; EGE University (Turkey) attempted to optimize a hydrogen-producing pathway in extremophilic bacteria; and the University of Edinburgh built an arsenic whole cell biosensor that gives a pH response which can be inexpensively read by methyl red or an electronic pH meter. The Edinburgh team's project was particularly interesting because they were able to design a complex system, test it in silico, and construct a simplified prototype for proof-of-concept.
To achieve high detection sensitivity and accuracy over a wide range of sample concentrations, the team used a plasmid-encoded arsenic resistance operon controlled by two repressor proteins, ArsD and ArsR, which respond to high and low concentrations of arsenate, respectively. An increase in pH is induced by urease (expressed from a hybrid promoter repressed by both lambda cI and LacI repressors), which breaks down urea to introduce ammonium ions; while a decrease in pH is achieved by lacZ, which encodes an enzyme that begins the fermentation of lactose to acid products.
In the presence of lactose (added to activate the system) and absence of arsenate, urease is induced to raise the pH. At low arsenate concentrations, the ArsR-repressed promoter is induced, leading to lambda cI repressor expression, which shuts down the production of urease to give near-neutral pH. Finally, at high arsenate concentrations, the ArsD-responsive promotor induces lacZ expression, lowering the pH.
The biosensor system was modeled in silico by eighteen ordinary differential equations, with gene expression reactions modeled by a Michaelis-Menten equation and the other reactions modeled by mass-action equations found in literature. This virtual experiment confirmed the system's functionality (at least in theory) and identified the most influential kinetic parameters (e.g. the degradation rate of ArsD).
A laboratory proof-of-concept construct was built by constructing a biobrick part containing the E. coli chrososomal ars promoter and a negatively autoregulated arsR gene, constructing another biobrick part based on the lacZ’ gene (encoding the N-terminus of lacZ, which complements a mutation found on the chromosome of laboratory E. coli), and combining the two parts. This prototype achieved a pH response to arsenate concentrations as low as 5 ppb arsenic, well below the World Health Organization's limit for acceptable drinking water (10 ppb).
http://www.igem2006.com/jamboree.htm http://parts2.mit.edu/wiki/index.php/Edinburgh_summary_page http://parts2.mit.edu/wiki/index.php/EdinburghModeling
Energy Applications
The need for alternative fuels has increased considerably in the recent years due to rising fossil fuel prices and worldwide attention towards environmental sustainability (hill06). While battery technology has steadily progressed, electrochemical energy storage still cannot (and may never) compete with liquid or gaseous fuels on energy density.
Among the promising candidates for alternative fuels are biofuels and hydrogen. Biofuels -- fuels produced from crop-based carbohydrates -- include bioethanol from fermentable sugars of corn or switchgrass, and biodiesel from plant oils (torney07). To improve biofuel production, numerous groups have employed recombinant and metabolic engineering to develop microorganisms that convert carbohydrates from a variety of feedstocks into biofuels. For example, a recombinant Saccharomyces yeast strain has been engineered to break down cellulose and ferment ethanol from xylose, a pentose commonly found in renewable lignocellulosic biomass such as waste paper. This was achieved by introducing genes from P. stipitis and S. cerevisiae for their xylose-fermenting ability, and displaying on the cell surface a cellooligosaccharide-degrading fusion protein from an A. aculeatus gene and α-agglutinin (katahira06). Commercial entities pursuing biofuel applications of synthetic biology include California startups Amyris Biotechnologies, LS9, and Synthetic Genomics.
Using a different synthetic biology approach to produce hydrogen gas, Zhang et al created a synthetic enzymatic pathway employing thirteen enzymes from rabbit muscle, spinach, and three strains of bacteria (S. cerevisiae, E. Coli, P. furiosus) to convert starch and water into hydrogen and carbon dioxide, as summarized in Table X (zhang07).
The resulting spontaneous (ΔG°= -48.9 kJ/mol), slightly endothermic (ΔH°= 595.6 kJ/mol) overall reaction
C6H10O5 (l) + H2O (l) ⇒ 12 H2 (g) + 6 CO2 (g)
occurred at 30°C under atmospheric pressure and yielded more hydrogen than the theoretical yield of biological hydrogen fermentation (12 versus 4 H2/glucose).
This synthetic enzymatic approach has important advantages over chemical catalysis for hydrogen production, including lower toxicity, milder reaction conditions, less stringent infrastructural requirements, and better economics from using starch as the feedstock. Furthermore, when compared with microbial approaches, synthetic enzymatic pathways have higher product yield, easier implementation and controls, no formation of by-products and cell mass, and broader reaction conditions (i.e. wider temperature and pH ranges). It is anticipated that cell-free synthetic enzymatic pathways will eventually replace chemical catalysis and microorganisms in the production of many commodity chemicals (zhang07).
This synthetic enzymatic approach has important advantages over chemical catalysis for hydrogen production, including lower toxicity, milder reaction conditions, less stringent infrastructural requirements, and better economics from using starch as the feedstock. Furthermore, when compared with microbial approaches, synthetic enzymatic pathways have higher product yield, easier implementation and controls, no formation of by-products and cell mass, and broader reaction conditions (i.e. wider temperature and pH ranges). It is anticipated that cell-free synthetic enzymatic pathways will eventually replace chemical catalysis and microorganisms in the production of many commodity chemicals (zhang07).
Some individuals--including Craig Venter, founder of The Institute for Genomic Research, the J. Craig Venter Institute for genomic research, and Synthetic Genomics--consider energy the most pressing issue on the planet today. Enormous challenges exist for producing high energy fuel inexpensively and without occupying too much fertile land. Fortunately, synthetic biology has recently matured enough to address some of these critical challenges. Given the existence of a variety of carbon-containing feedstock, metabolic and enzymatic pathways, genetically modifiable organisms, and R&D funding sources, we can expect a significant amount of biological innovation to come from the energy sector within the next decade.
- Torney F, Moeller L, Scarpa A, and Wang K. Genetic engineering approaches to improve bioethanol production from maize. Curr Opin Biotechnol. 2007 Jun;18(3):193-9. DOI:10.1016/j.copbio.2007.03.006 |
- Katahira S, Mizuike A, Fukuda H, and Kondo A. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol. 2006 Oct;72(6):1136-43. DOI:10.1007/s00253-006-0402-x |
- Hill J, Nelson E, Tilman D, Polasky S, and Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci U S A. 2006 Jul 25;103(30):11206-10. DOI:10.1073/pnas.0604600103 |
- Zhang YH, Evans BR, Mielenz JR, Hopkins RC, and Adams MW. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS One. 2007 May 23;2(5):e456. DOI:10.1371/journal.pone.0000456 |
http://www.msnbc.msn.com/id/18882837/site/newsweek/
How 'Synthetic' is Environmental Synthetic Biology?
The current state of knowledge in biology has enabled researchers to work principally within one level of the abstraction hierarchy to engineer novel organisms. For example, a biological input-output device can be created by taking an output part from one organism (e.g. the GFP gene from jellyfish [Nature425]), and input part from another (e.g. an olfactory receptor from a mammalian cell [Pajot-Augy03]), and integrating those parts into a chassis of choice (e.g. yeast cells). This sort of genetic plug-and-play can in some cases be done without fully understanding the operational mechanisms of all the parts.
However, the lack of standardization of biological parts and devices has impeded research progress by requiring researchers to devote a significant portion of their efforts to interface engineering -- that is, assembling the many parts and getting them to function together predictably. Standardization would reduce costly inefficiencies by simplifying both the design and construction stages of bioengineering and minimizing the amount of experimental guesswork needed.
Furthermore, as bioconstruction technologies advance, synthetic biology research activities can be expected to progress at higher rates and become much more widespread. While researchers today can purchase made-to-order DNA cheaply, most of the necessary bioengineering steps (e.g. making fusion proteins,XXXXXXXXXXXX) must still be done laboriously by hand. Decoupling design and construction in bioengineering would allow individuals to innovate without having to work in expensive wet lab facilities at all times. Just as online machine shop services have enabled individuals to order custom-designed parts to create innovative products from their home, the further decoupling of biological design and construction (e.g. providing the ability to order custom-designed organisms rather than just fragments its DNA) will allow synthetic biologist to create and experiment with useful biological systems at lower costs. Perhaps more importantly, the decoupling would enable a greater number of individuals (of various levels of experience) to utilize synthetic biology for solving a variety of real world problems.
In the following sections, the current state of environmental synthetic biology is summarized. It shall be apparent that most of the work involve creating whole cell sensors that contain a reporter (e.g. coding for GFP or luciferase) and a promoter inducible by environmental toxins. These projects can be considered device-level engineering, as the resulting product is usually a simple input-output device. Examples of system-level engineering for environmental applications is only starting to appear...
References
1. Abd-El-Haleem D, Ripp S, Scott C, Sayler GS (2002) A luxCDABE-based bioluminescent bioreporter for the detection of phenol. Journal of Industrial Microbiology & Biotechnology 29: 233-237.
2. Aldor AS, Keasling JD (2003) Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Current Opinion in Biotechnology 14: 475-483.
3. Bae W, Mehra RK, Mulchandani A, Chen W (2001) Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Applied and Environmental Microbiology 67: 5335-5338.
4. Belkin S (2003) Microbial whole-cell sensing systems of environmental pollutants. Current Opinion in Microbiology 6: 206-212.
5. Benner SA, Sismour AM (2005) Synthetic biology. Nature Reviews Genetics 6: 533-543.
6. Daunert S, Barrett G, Feliciano JS, Shetty RS, Shrestha S, et al. (2000) Genetically engineered whale-cell sensing systems: Coupling biological recognition with reporter genes. Chemical Reviews 100: 2705-2738.
7. Drubin DA, Way JC, Silver PA (2007) Designing biological systems. Genes & Development 21: 242-254.
8. Endy D (2005) Foundations for engineering biology. Nature 438: 449-453.
9. Gu MB, Min J, Kim EJ (2002) Toxicity monitoring and classification of endocrine disrupting chemicals (EDCs) using recombinant bioluminescent bacteria. Chemosphere 46: 289-294.
10. Harms H, Wells MC, van der Meer JR (2006) Whole-cell living biosensors - are they ready for environmental application? Applied Microbiology and Biotechnology 70: 273-280.
11. Hever N, Belkin S (2006) A dual-color bacterial reporter strain for the detection of toxic and genotoxic effects. Engineering in Life Sciences 6: 319-323.
12. Hezayen FF, Steinbuchel A, Rehm BHA (2002) Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56. Archives of Biochemistry and Biophysics 403: 284-291.
13. Hill J, Nelson E, Tilman D, Polasky S, Tiffany D (2006) Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proceedings of the National Academy of Sciences of the United States of America 103: 11206-11210.
14. Katahira S, Mizuike A, Fukuda H, Kondo A (2006) Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Applied Microbiology and Biotechnology 72: 1136-1143.
15. Keane A, Phoenix P, Ghoshal S, Lau PCK (2002) Exposing culprit organic pollutants: A review. Journal of Microbiological Methods 49: 103-119.
16. Kostrzynska M, Leung KT, Lee H, Trevors JT (2002) Green fluorescent protein-based biosensor for detecting SOS-inducing activity of genotoxic compounds. Journal of Microbiological Methods 48: 43-51.
17. Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, et al. (2005) Engineering Escherichia coli to see light - These smart bacteria 'photograph' a light pattern as a high-definition chemical image. Nature 438: 441-442.
18. Pleiss J (2006) The promise of synthetic biology. Applied Microbiology and Biotechnology 73: 735-739.
19. Shao CY, Howe CJ, Porter AJR, Glover LA (2002) Novel cyanobacterial biosensor for detection of herbicides. Applied and Environmental Microbiology 68: 5026-5033.
20. Sorensen SJ, Burmolle M, Hansen LH (2006) Making bio-sense of toxicity: new developments in whole-cell biosensors. Current Opinion in Biotechnology 17: 11-16.
21. Steinbuchel A, Fuchtenbusch B (1998) Bacterial and other biological systems for polyester production. Trends in Biotechnology 16: 419-427.
22. Stocker J, Balluch D, Gsell M, Harms H, Feliciano J, et al. (2003) Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environmental Science & Technology 37: 4743-4750.
23. Suriyamongkol P, Weselake R, Narine S, Moloney M, Shah S (2007) Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants - A review. Biotechnology Advances 25: 148-175.
24. Vandyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, et al. (1994) RAPID AND SENSITIVE POLLUTANT DETECTION BY INDUCTION OF HEAT-SHOCK GENE-BIOLUMINESCENCE GENE FUSIONS. Applied and Environmental Microbiology 60: 1414-1420.
25. Vollmer AC, Belkin S, Smulski DR, VanDyk TK, LaRossa RA (1997) Detection of DNA damage by use of Escherichia coli carrying recA'-lux, uvrA'-lux, or alkA'-lux reporter plasmids. Applied and Environmental Microbiology 63: 2566-2571.
26. Ward PG, Goff M, Donner M, Kaminsky W, O'Connor KE (2006) A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environmental Science & Technology 40: 2433-2437.
27. Wells M, Gosch M, Harms H, van der Meer JR (2005) Response characteristics of arsenic-sensitive bioreporters expressing the gfp reporter gene. Microchimica Acta 151: 209-216.
28. Wells M, Gosch M, Rigler R, Harms H, Lasser T, et al. (2005) Ultrasensitive reporter protein detection in genetically engineered bacteria. Analytical Chemistry 77: 2683-2689.
29. Yagi K (2007) Applications of whole-cell bacterial sensors in biotechnology and environmental science. Applied Microbiology and Biotechnology 73: 1251-1258.
References to be added manually
Curr Opin Biotechnol. 2007 Jun;18(3):193-9. Epub 2007 Mar 30. Genetic engineering approaches to improve bioethanol production from maize. Torney F, Moeller L, Scarpa A, Wang K. Center for Plant Transformation, Plant Science Institute and Department of Agronomy, Iowa State University, Ames, Iowa 50011, USA.
PLoS ONE. 2007 May 23;2:e456.Click here to read Click here to read Links
High-yield hydrogen production from starch and water by a synthetic enzymatic pathway.
Zhang YH, Evans BR, Mielenz JR, Hopkins RC, Adams MW.
Biological Systems Engineering Department, Virginia Tech, Blacksburg, Virginia, United States of America.
Making It Happen
June 4, 2007
Craig Venter galvanized the Human Genome Project. Can he do it for synthetic biology?
By Barrett Sheridan
Newsweek International
"Modeling the Arsenic Biosensor System" by the Edinburgh iGEM team
Non-relevant
Calculating relative centrifugal force (RCF)
Relative centrifugal force is the measurement of the force applied to a sample within a centrifuge. This can be calculated from the speed (RPM) and the rotational radius (cm) using the following calculation.
- g = RCF = 0.00001118 × r × N2
where
- g = Relative centrifuge force
- r = rotational radius (centimetre, cm)
- N = rotating speed (revolutions per minute, r/min)
Links
Title:
Selective Fractionation of Nanowire Diameter by Centrifugation
Authors:
Trammell, T. E.
Publication:
American Physical Society, APS March Meeting, March 21-25, 2005, abstract #V25.005
Publication Date:
03/2005
http://www.coleparmer.com/techinfo/techinfo.asp?htmlfile=basic-centrifugation.htm&ID=30
http://homepages.gac.edu/~cellab/appds/appd-f.html
Project
(Glu-Cys)20Gly "EC20" binds heavy metals such as mercury -- express it on phage!
Bind p3 of phage to magnetic bead -- use bead to remove mercury from water