User:Student 66/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
'''Introduction''' | '''Introduction''' | ||
The purpose of the following experiment was to identify and characterize the colonies and cells of bacterial species collected in Transect #3 after having left the serially diluted agar plates to grow over the course of a week. We were able to make our observations with the aid of a wet mount to clearly see the behavior of the cells under the microscope. Following that, we used the technique called Gram stain to observe the cell walls and determine the amount of peptidoglycan in the bacteria. Our hypothesis was that the the bacteria found on the regular agar plates [tet(-)] would obtain a wider variety and amount of bacteria than the bacteria observed on the agar plates with tetracycline [tet(+)]. | The purpose of the following experiment was to identify and characterize the colonies and cells of bacterial species collected in Transect #3 after having left the serially diluted agar plates to grow over the course of a week. We were able to make our observations with the aid of a wet mount to clearly see the behavior of the cells under the microscope. Following that, we used the technique called Gram stain to observe the cell walls and determine the amount of peptidoglycan in the bacteria. Our hypothesis was that the the bacteria found on the regular agar plates [tet(-)] would obtain a wider variety and amount of bacteria than the bacteria observed on the agar plates with tetracycline [tet(+)].* | ||
What we found was that our serial dilutions (from a range of 10^-3 to 10^-9 - odd numbers only) supported our hypothesis. The group without tetracycline displayed more colonies: 10^-3=880 10^-5=213 10^-7=150 and 10^-9=80. We can make the assumption that the greater the dilution number the more resistant it is to bacterial and fungal growth. | |||
Our second group with tetracycline displayed less colonies: 10^-3=28 10^-5=1 10^-7=0 and 10^-9=0. We can make the assumption that the lower the tetracycline dilution, the higher the chance of forming colonies on agar plates. | |||
When observing the Hay Infusion, we found that there were a couple changes in smell and appearance. The color of our Hay Infusion had changed from a dark shade of green to a light. There was also a lack of water compared to when we first observed our Hay Infusion. The smell was just as bad as before but less pungent - we believe that this is due to the breaking down of microorganisms by the bacteria. | When observing the Hay Infusion, we found that there were a couple changes in smell and appearance. The color of our Hay Infusion had changed from a dark shade of green to a light. There was also a lack of water compared to when we first observed our Hay Infusion. The smell was just as bad as before but less pungent - we believe that this is due to the breaking down of microorganisms by the bacteria. | ||
'''* Tetracyclines are inexpensive antibiotics, which have been used extensively in the prophlylaxis and therapy of human and animal infections and also at subtherapeutic levels in animal feed as growth promoters. Tetracycline are broad-spectrum agents, exhibiting activity against a wide range of gram-positive and gram-negative bacteria, atypical organisms such as chlamydiae, mycoplasmas, and rickettsiae, and protozoan parasites.Tetracyclines inhibit bacterial protein synthesis by preventing the association of aminoacyl-tRNA with the bacterial ribosome.''' | |||
''Chopra,I. June 2001 "Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance". Journal of Microbiology and Molecular Biology Review. Volume 65(2).'' | |||
'''Materials and Methods''' | '''Materials and Methods''' | ||
1. Collect the 8 agar dishes that were set aside the week before. Observe colony morphology and | 1. Collect the 8 agar dishes that were set aside the week before. Observe colony morphology and take note of any changes. | ||
2. Retrieve 2 samples from (+) and 2 samples from (-) and place them on glass slides. | |||
3. Begin the Gram Stain process by evaporating excess water with the air of a Bunsen burner. | |||
4. Continue the Gram stain process from the sheets posted on the lab tables. You will be using Crystal Violet, Iodine, Alcohol, and then Safranin Stain, and rinsing with deionized water in between steps. | |||
5. Let the glass plate dry completely and proceed to observe under the microscope. Record whether or not are Gram positive or negative by basing it off the color of the peptidoglycan. | |||
6. Prepare wet mounts for the chosen samples from the agar dishes and observe and record your observations of the behavior of bacterial species. | |||
7. Take samples of bacteria, (-) and (+), and place it into two PCR tubes for genetic analysis. | |||
'''Results''' | '''Results''' | ||
| Line 24: | Line 36: | ||
''Table 1: Serial dilution results table'' | ''Table 1: Serial dilution results table'' | ||
[[Image:ETETBAM.png]] | |||
''Table 2: Serial Diluted Agar Plates'' | |||
[[Image:EPLATEBAM.png]] | |||
''Table 3: Gram stains: Plates E7, E9, E5, E3'' | |||
'''Conclusion''' | |||
Our hypothesis appeared to be correct. There was a more diverse and abundant amount of bacterial growth on the agar plates that did not obtain tetracycline [tet(-)]. | |||
-MR | -MR | ||
---- | ---- | ||
Revision as of 12:17, 11 February 2016
Week 3: Characterizing Colonies Using Gram Stains and Wet Mounts
Introduction The purpose of the following experiment was to identify and characterize the colonies and cells of bacterial species collected in Transect #3 after having left the serially diluted agar plates to grow over the course of a week. We were able to make our observations with the aid of a wet mount to clearly see the behavior of the cells under the microscope. Following that, we used the technique called Gram stain to observe the cell walls and determine the amount of peptidoglycan in the bacteria. Our hypothesis was that the the bacteria found on the regular agar plates [tet(-)] would obtain a wider variety and amount of bacteria than the bacteria observed on the agar plates with tetracycline [tet(+)].*
What we found was that our serial dilutions (from a range of 10^-3 to 10^-9 - odd numbers only) supported our hypothesis. The group without tetracycline displayed more colonies: 10^-3=880 10^-5=213 10^-7=150 and 10^-9=80. We can make the assumption that the greater the dilution number the more resistant it is to bacterial and fungal growth.
Our second group with tetracycline displayed less colonies: 10^-3=28 10^-5=1 10^-7=0 and 10^-9=0. We can make the assumption that the lower the tetracycline dilution, the higher the chance of forming colonies on agar plates.
When observing the Hay Infusion, we found that there were a couple changes in smell and appearance. The color of our Hay Infusion had changed from a dark shade of green to a light. There was also a lack of water compared to when we first observed our Hay Infusion. The smell was just as bad as before but less pungent - we believe that this is due to the breaking down of microorganisms by the bacteria.
* Tetracyclines are inexpensive antibiotics, which have been used extensively in the prophlylaxis and therapy of human and animal infections and also at subtherapeutic levels in animal feed as growth promoters. Tetracycline are broad-spectrum agents, exhibiting activity against a wide range of gram-positive and gram-negative bacteria, atypical organisms such as chlamydiae, mycoplasmas, and rickettsiae, and protozoan parasites.Tetracyclines inhibit bacterial protein synthesis by preventing the association of aminoacyl-tRNA with the bacterial ribosome.
Chopra,I. June 2001 "Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance". Journal of Microbiology and Molecular Biology Review. Volume 65(2).
Materials and Methods
1. Collect the 8 agar dishes that were set aside the week before. Observe colony morphology and take note of any changes.
2. Retrieve 2 samples from (+) and 2 samples from (-) and place them on glass slides.
3. Begin the Gram Stain process by evaporating excess water with the air of a Bunsen burner.
4. Continue the Gram stain process from the sheets posted on the lab tables. You will be using Crystal Violet, Iodine, Alcohol, and then Safranin Stain, and rinsing with deionized water in between steps.
5. Let the glass plate dry completely and proceed to observe under the microscope. Record whether or not are Gram positive or negative by basing it off the color of the peptidoglycan.
6. Prepare wet mounts for the chosen samples from the agar dishes and observe and record your observations of the behavior of bacterial species.
7. Take samples of bacteria, (-) and (+), and place it into two PCR tubes for genetic analysis.
Results
Table 1: Serial dilution results table
Table 2: Serial Diluted Agar Plates
Table 3: Gram stains: Plates E7, E9, E5, E3
Conclusion
Our hypothesis appeared to be correct. There was a more diverse and abundant amount of bacterial growth on the agar plates that did not obtain tetracycline [tet(-)].
-MR
Week 2: Hay Infusion
Introduction The purpose of the following experiment to observe the unicellular eukaryotic organisms growing on the top, middle, and bottom of Transect #3's Hay Infusion Culture. We used a dichotomous key to identify the protists and algae found in Transect #3. Our hypothesis would be that the different niches (top, middle, bottom) would have different protists and algae due to the different abiotic and biotic components in each layer.
Materials and Methods
1. Observe the Hay Infusion Culture. Make any notes on the appearance, smell, and color.
2. Identify the different layers of the Hay Infusion Culture and describe each.
3. Observe a sample from each niche.
4. Measure each organism and identify each using a dichotomous key.
5. Mix the Hay Infusion Culture jar by swirling it around with the lid on.
6. Collect and label four tubes of 10mLs sterile broth with 10^-2, 10^-4,10^-6,10^-8. Also collect a micropippetor and set is at 100micro Liters.
7. Collect four nutrient agar and four nutrient agar plus tetracycline plates. Label the plates with tetracycline with "tet" - always on the side of the plate.
8. Add 100 micro Liters from the culture to the 10mLs of broth in the tube labeled 10^-2. Swirl the inoculated tube throughly.
9. Repeat this process twice more to make the 10^-6 and 10^-8 dilutions.
10. Pipette 100 micro Liters from the 10^-2 tube onto the nutrient agar plate labeled 10^-3.
11. Repeat this process on the +tet plate labeled 10^-3.
12. Repeat this procedure with the 10^-4 dilution on the 10^-5 plates, 10^-6 dilution on the 10^-7 plates and the 10^-8 dilution on the 10^-9 plates.
13. Set the agar plates aside (side up) onto a rack. Leave them there at room temperature for a week.
Results
Observations of Hay Infusion Culture

Figure 1: Transect #3 Jar
Figure 2: Transect #3 Jar from above
1. The smell was pungent, like sewer. Almost a mold-like smell. Murky in appearance. The water was a light brown/green.
2. There was life on top of the liquid, as seen by the mold film on the surface of the water.
Niches of our jar:'
Figure 3: Jar Niches - Three distinct niches including: top, middle, bottom
4. Protists and algae present
On the top layer: Gonium, pandorina, colpidium, volvox
Middle layer: Volvox, paramecium bursaria, spirostomum, eugirna
Bottom layer: Blepharisma, didinium cyst, chilomonas sp,, spirostomum, pandorina colony
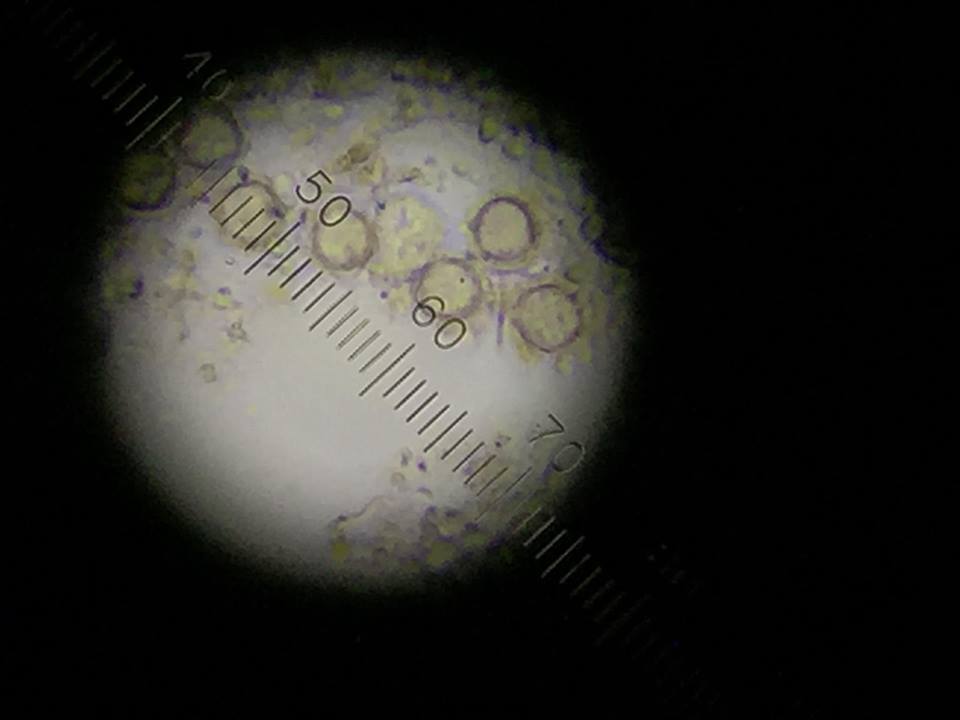 Figure 4: Gonium found top niche.
Figure 4: Gonium found top niche.
The following organism was found in the top layer of our jar. Observed was the gonium (colony up to 90 um).
 Figure 5: Volvox found middle niche.
Figure 5: Volvox found middle niche.
The following organism was found in the middle layer of our jar. Observed was the volvox (350-500 um).
 Figure 6: Didinium cyst found on bottom layer.
Figure 6: Didinium cyst found on bottom layer.
The following organism was found in the bottom layer of our jar. Observed was the didinium cyst.
6. None of the following above were motile.
7. If the Hay Infusion Culture "grew" for another two months, it is likely that the protozoans would consume the algal organisms. It is likely that mold would also begin to grow.
Serial Dilution:
Figure 7: Serial Dilution Process
Conclusion
Our hypothesis appeared to be correct. There were different protists and algae found in the top, middle, and bottom niches. This provides insight that certain organisms thrive in different niches of abiotic and biotic features.
-MR
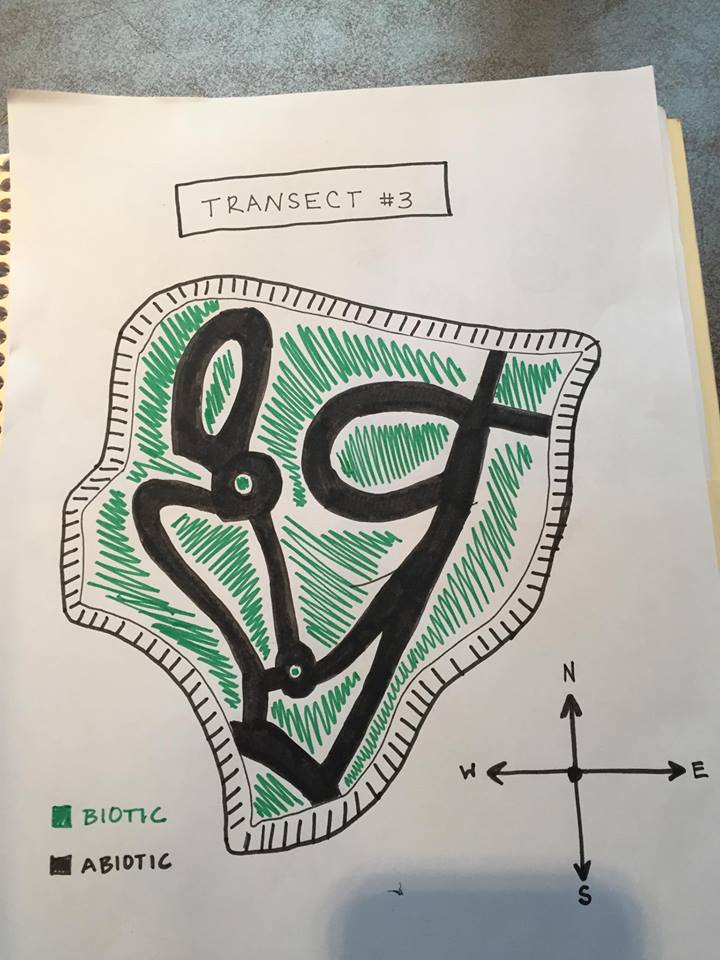
Transect #3
Week 1: Transect Observation
Location:
Aerial view of Transect showing Abiotic and Biotic areas:
Transect #3 sits between Bender Arena and Hughes Residence Hall. The transect lies just above the amphitheater and before the road. The area consists of a few benches, light poles, garbage can, and a concrete pathways winding about the garden. The ground is relatively flat only to be interrupted with a staircase. The soil is dark with mulch and dead leaves on top of it. There are a variety of trees and plants assorted throughout the transect as well.
Abiotic features: 1. Metal benches 2. Mulch 3. Rocks 4. Dirt 5. Lamp poles
Biotic features: 1.Leafy plants 2. Leaves 3. Weeds 4. Tall trees 5. Small insects
Madeline_B._Rohrbacher