User:Saroj Pandey/Notebook/SNP PCR optimization/2014/09/18: Difference between revisions
Saroj Pandey (talk | contribs) |
Saroj Pandey (talk | contribs) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
| colspan="2"| | | colspan="2"| | ||
<!-- ##### DO NOT edit above this line unless you know what you are doing. ##### --> | <!-- ##### DO NOT edit above this line unless you know what you are doing. ##### --> | ||
== | ==PCR for gDNA amplification== | ||
'''Primers''' | |||
Product Size: 992 bp Pair Any: 6.0 Pair End: 2.0 GC: 45.3% | |||
TAS2R38_gaF; ATCCGTGATGCTGTGCTATG | |||
Length: | Length: 20 bp Tm: 59.7 °C GC: 50.0 % ANY: 4.0 SELF: 0.0 | ||
TAS2R38_gaR; GCATCCCAGAAGAAACCAGA | |||
Length: 20 bp Tm: 60.2 °C GC: 50.0 % ANY: 2.0 SELF: 0.0 | |||
'''PCR 1''' | |||
• A PCR was run using Phusion DNA polymerase | |||
• Genomic DNA extracted from taster and non-taster were used as templates | |||
• Water was used in place of template in the blank | |||
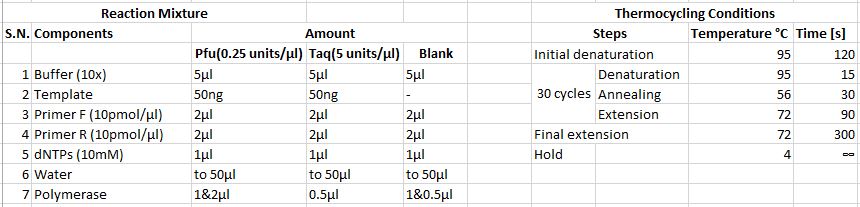
• Following reaction mixtures and thermocycling conditions were applied | |||
[[Image: gDNA_PCR1.JPG]] | |||
'''Agarose gel electrophoresis''' | |||
Gel preparation | |||
1. 0.7% agarose gel was prepared, for which 0.7g agarose was mixed in 100 ml water | |||
2. The mixture was heated at 600 °C for 90 seconds to completely dissolve the agarose | |||
3. 10µl gel red was added to the solution and mixed thoroughly | |||
4. Casting apparatus and comb was prepared | |||
5. The agarose solution was poured into the casting apparatus when lukewarm | |||
Sample Preparation | |||
loading dye [30% (v/v) glycerol + 0.25% (w/v) brophenol blue] | |||
• Ladder: 5µl water + 2µl loading dye + 2µl DNA ladder | |||
• Sample: 5µl water + 2µl loading dye + 5µl PCR product | |||
Sample loading and electrophoresis | |||
1. Approximately 450 ml of TAE buffer was poured into the electrophoretic chamber | |||
2. After the gel was set, it was transferred to the electrophoretic chamber and the comb was removed slowly without disrupting the sample wells | |||
3. Samples and DNA ladders were loaded into corresponding wells | |||
4. The electrophoretic chamber was closed and was connected to power supply | |||
5. Electrophoresis was run at 120V for 30 minutes | |||
6. The gel was observed under UV light | |||
[[Image:Electrophoresis_GA_04.09.jpg|right|thumb|350px|Electrophoresis]] | |||
Observations: | |||
• 100 bp DNA ladder and first few samples could not be observed in agarose gel under UV light | |||
• 1 kb DNA ladder was fluffy and diffused. Bands were not distinct. | |||
• All the PCR products (taster, non-taster and blank) showed diffused bands below 250bp | |||
Conclusions: | |||
• Required segment of the genomic DNA was not amplified | |||
• The bands seen could be primer dimers or other unspecific product | |||
'''PCR 2''' | '''PCR 2''' | ||
[[Image: | • As the desired region of the gDNA was not amplified, PCR was performed again | ||
[[Image: | |||
• This time, Taq DNA polymerase was also used in addition to the Phusion DNA polymerase | |||
• The concentration of Phusion DNA polymerase was reduced and two different volumes were used for each template | |||
• Annealing temperature was also reduced | |||
[[Image: gDNA_PCR2.JPG]] | |||
[[Image:Electrophoresis_GA_05.09.jpg|right|thumb|350px|Electrophoresis]] | |||
[[Image:Electrophoresis_GA_15.09.jpg|right|thumb|350px|Electrophoresis]] | |||
'''Agarose gel electrophoresis''' | |||
• Similar procedure was adopted as described above | |||
Observations: | |||
• 100 bp DNA ladder appears like a smear. No bands could be detected. | |||
• Although the bands could be identified in 1 kb DNA ladder, they are diffused and unclear. | |||
• On repeated PCR with less amount of Phusion polymerase, no desired product (992 bp) was observed | |||
• There was an expected band at 1kb in PCR products with Taq DNA polymerase for both the taster and the non-taster | |||
• All samples and blank with Taq DNA polymerase and a taster sample with 0.5 units of Phusion DNA polymerase showed bands below 250 bp | |||
• All of the DNA bands observed were diffused and/or elongated | |||
Concusion: | |||
• One reason for unclear gel picture could be the preparation of gel itself. The gel was prepared only in water and not in buffer. | |||
• The reason for unspecific products could be because of the use of high amount of DNA polymerases for PCR reaction | |||
'''Agarose gel electrophoresis (repeated)''' | |||
• Although it was observed that desired segment of the genomic DNAs were amplified, the gel picture was unclear. So electrophoresis was repeated for better result. | |||
• This time, agarose gel was prepared in TAE buffer instead of water. Every other procedure was the same. | |||
Observations: | |||
• DNA ladders are seen with clear and distinct bands | |||
• As previously observed, Taq DNA polymerase was able to amplify the desired DNA segment | |||
• Unspecific products were observed in some samples | |||
Conclusions: | |||
• Use of TAE buffer instead of water greatly improves the quality of gel for the separation of DNA | |||
'''DNA concentration''' | |||
DNA concentrations in the PCR products with desired DNA segment were measured | |||
• Taster: 520.9 ng/dl [260:280 = 1.79] | |||
• Non-taster: 406.7 ng/dl [260:280 = 1.83] | |||
These PCR products were sent for sequencing | |||
Revision as of 15:59, 21 September 2014
| <html><img src="/images/9/94/Report.png" border="0" /></html> Main project page <html><img src="/images/c/c3/Resultset_previous.png" border="0" /></html>Previous entry<html> </html>Next entry<html><img src="/images/5/5c/Resultset_next.png" border="0" /></html> | |
PCR for gDNA amplificationPrimers Product Size: 992 bp Pair Any: 6.0 Pair End: 2.0 GC: 45.3% TAS2R38_gaF; ATCCGTGATGCTGTGCTATG Length: 20 bp Tm: 59.7 °C GC: 50.0 % ANY: 4.0 SELF: 0.0 TAS2R38_gaR; GCATCCCAGAAGAAACCAGA Length: 20 bp Tm: 60.2 °C GC: 50.0 % ANY: 2.0 SELF: 0.0
• A PCR was run using Phusion DNA polymerase • Genomic DNA extracted from taster and non-taster were used as templates • Water was used in place of template in the blank • Following reaction mixtures and thermocycling conditions were applied
Agarose gel electrophoresis Gel preparation 1. 0.7% agarose gel was prepared, for which 0.7g agarose was mixed in 100 ml water 2. The mixture was heated at 600 °C for 90 seconds to completely dissolve the agarose 3. 10µl gel red was added to the solution and mixed thoroughly 4. Casting apparatus and comb was prepared 5. The agarose solution was poured into the casting apparatus when lukewarm
loading dye [30% (v/v) glycerol + 0.25% (w/v) brophenol blue] • Ladder: 5µl water + 2µl loading dye + 2µl DNA ladder • Sample: 5µl water + 2µl loading dye + 5µl PCR product
1. Approximately 450 ml of TAE buffer was poured into the electrophoretic chamber 2. After the gel was set, it was transferred to the electrophoretic chamber and the comb was removed slowly without disrupting the sample wells 3. Samples and DNA ladders were loaded into corresponding wells 4. The electrophoretic chamber was closed and was connected to power supply 5. Electrophoresis was run at 120V for 30 minutes 6. The gel was observed under UV light  Observations: • 100 bp DNA ladder and first few samples could not be observed in agarose gel under UV light • 1 kb DNA ladder was fluffy and diffused. Bands were not distinct. • All the PCR products (taster, non-taster and blank) showed diffused bands below 250bp
• Required segment of the genomic DNA was not amplified • The bands seen could be primer dimers or other unspecific product
• As the desired region of the gDNA was not amplified, PCR was performed again • This time, Taq DNA polymerase was also used in addition to the Phusion DNA polymerase • The concentration of Phusion DNA polymerase was reduced and two different volumes were used for each template • Annealing temperature was also reduced   Agarose gel electrophoresis • Similar procedure was adopted as described above
• 100 bp DNA ladder appears like a smear. No bands could be detected. • Although the bands could be identified in 1 kb DNA ladder, they are diffused and unclear. • On repeated PCR with less amount of Phusion polymerase, no desired product (992 bp) was observed • There was an expected band at 1kb in PCR products with Taq DNA polymerase for both the taster and the non-taster • All samples and blank with Taq DNA polymerase and a taster sample with 0.5 units of Phusion DNA polymerase showed bands below 250 bp • All of the DNA bands observed were diffused and/or elongated
• One reason for unclear gel picture could be the preparation of gel itself. The gel was prepared only in water and not in buffer. • The reason for unspecific products could be because of the use of high amount of DNA polymerases for PCR reaction
• Although it was observed that desired segment of the genomic DNAs were amplified, the gel picture was unclear. So electrophoresis was repeated for better result. • This time, agarose gel was prepared in TAE buffer instead of water. Every other procedure was the same.
• DNA ladders are seen with clear and distinct bands • As previously observed, Taq DNA polymerase was able to amplify the desired DNA segment • Unspecific products were observed in some samples
• Use of TAE buffer instead of water greatly improves the quality of gel for the separation of DNA
DNA concentrations in the PCR products with desired DNA segment were measured • Taster: 520.9 ng/dl [260:280 = 1.79] • Non-taster: 406.7 ng/dl [260:280 = 1.83] These PCR products were sent for sequencing
| |