User:Rebeca Rodriguez/Notebook/Chem 471/2015/11/11: Difference between revisions
(Autocreate 2015/11/11 Entry for User:Rebeca_Rodriguez/Notebook/Chem_471) |
No edit summary |
||
| Line 1: | Line 1: | ||
{|{{table}} width="800" | {|{{table}} width="800" | ||
|- | |- | ||
|style="background-color: #EEE"|[[Image: | |style="background-color: #EEE"|[[Image:owwnotebook_icon.png|128px]]<span style="font-size:22px;"> Project name</span> | ||
|style="background-color: #F2F2F2" align="center"|<html><img src="/images/9/94/Report.png" border="0" /></html> [[{{#sub:{{FULLPAGENAME}}|0|-11}}|Main project page]]<br />{{#if:{{#lnpreventry:{{FULLPAGENAME}}}}|<html><img src="/images/c/c3/Resultset_previous.png" border="0" /></html>[[{{#lnpreventry:{{FULLPAGENAME}}}}{{!}}Previous entry]]<html> </html>}}{{#if:{{#lnnextentry:{{FULLPAGENAME}}}}|[[{{#lnnextentry:{{FULLPAGENAME}}}}{{!}}Next entry]]<html><img src="/images/5/5c/Resultset_next.png" border="0" /></html>}} | |style="background-color: #F2F2F2" align="center"|<html><img src="/images/9/94/Report.png" border="0" /></html> [[{{#sub:{{FULLPAGENAME}}|0|-11}}|Main project page]]<br />{{#if:{{#lnpreventry:{{FULLPAGENAME}}}}|<html><img src="/images/c/c3/Resultset_previous.png" border="0" /></html>[[{{#lnpreventry:{{FULLPAGENAME}}}}{{!}}Previous entry]]<html> </html>}}{{#if:{{#lnnextentry:{{FULLPAGENAME}}}}|[[{{#lnnextentry:{{FULLPAGENAME}}}}{{!}}Next entry]]<html><img src="/images/5/5c/Resultset_next.png" border="0" /></html>}} | ||
|- | |- | ||
| colspan="2"| | | colspan="2"| | ||
<!-- ##### DO NOT edit above this line unless you know what you are doing. ##### --> | <!-- ##### DO NOT edit above this line unless you know what you are doing. ##### --> | ||
==Repeat of Bradford for 10 nm Proteinase K== | |||
Repeat Bradford analysis of Degradation of AuNP Fibers using 10 nm Proteinase K. | |||
==Bradford Analysis of Protease Degradation== | |||
The general procedure today was taken from [[User:Matt_Hartings/Notebook/AU_Biomaterials_Design_Lab/2015/10/06#Bradford_Analysis_of_Protease_Degradation|Dr. Hartings's notebook]]. | |||
All samples will be incubated in eppendorf tubes, which will be placed in the 37°C water bath on the shaker. | |||
Samples should all contain the same concentration of materials. | |||
You should make samples for measurements at 10 minutes, 15 minutes, 30 minutes, 45 minutes, 1 hr, 1.5 hr, 2 hr. (Note, this is for the first time you do this. As you get better, I will expect you to get more measurements done). | |||
# Protease Prep | |||
## Place the 1mL of Tris/CaCl2 buffer in the pre-measured proteinase k '''(0.000111 g)''' sample and record the concentration '''(3.84·10<sup>-5</sup> M, second dilution: 3.84·10<sup>-7</sup> M)''' | |||
# Sample Prep | |||
## To a fiber sample tube (that has been centrifuge for 10 minutes at 300 RPM and had the supernatant removed) add the appropriate amounts of buffer and proteinase k '''(26.0 μL Proteinase K, 974.0 μL Buffer)'''. | |||
### The total volume should be 1mL | |||
### The final proteinase K concentration should be 10nM | |||
### Vortex the sample to disperse the fibers | |||
# Blank Prep (you will have one blank to match each sample) | |||
## To a clean eppendorf tube add the appropriate amounts of buffer and proteinase k '''(26.0 μL Proteinase K, 974.0 μL Buffer)'''. | |||
### The total volume should be 1mL | |||
### The final proteinase k concentration should be 10nM | |||
# Incubate Sample and Blanks | |||
## Place the tubes (for both the Blanks and Samples) in the shaker with the 37°C water bath | |||
# Measurement | |||
## Remove the tubes (the sample and a corresponding blank) from the water bath at the appropriate time | |||
## For the case of a sample with fibers, centrifuge the sample for 1 min to pull any fibers to the bottom of the tube | |||
## To a plastic cuvette add | |||
### 600 uL of pre-mixed Bradford dilution | |||
### 750 uL of sample | |||
### 1650 uL of buffer | |||
## Record the UV-Vis spectrum from 400-800 nm | |||
# Using the Bradford standardization curve and your data from the 0 fiber sample, determine the concentration of released protein in your solution. | |||
==Data== | |||
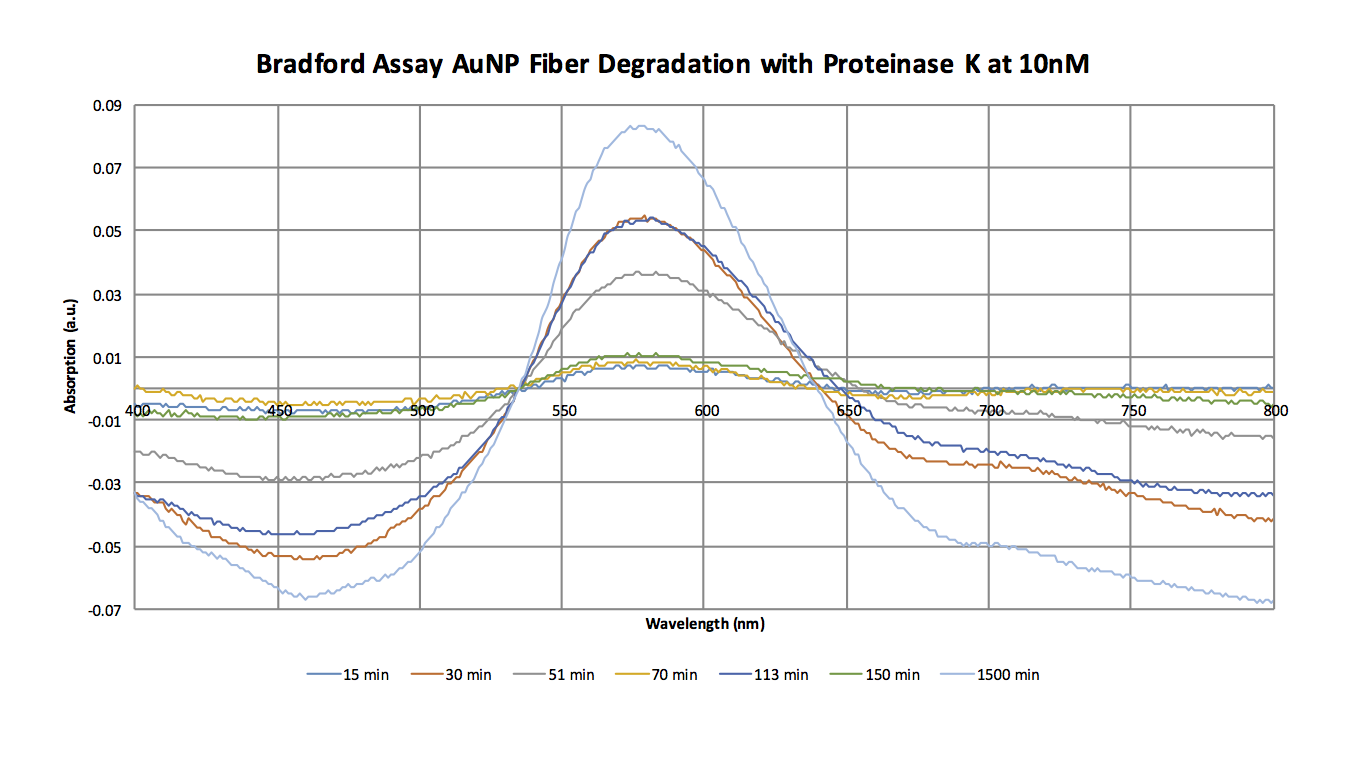
The spectrum for the overall blank with no protease or fiber sample was subtracted from all other spectra (blank and sample spectra). The spectra were further corrected by subtracting the absorbance at the isosbestic point. This data was redone from November 4th, because of the variation. | |||
[[Image:Bradford_10nM_Spectra_Nov_14.png]] | |||
Revision as of 16:32, 23 November 2015
| <html><img src="/images/9/94/Report.png" border="0" /></html> Main project page <html><img src="/images/c/c3/Resultset_previous.png" border="0" /></html>Previous entry<html> </html>Next entry<html><img src="/images/5/5c/Resultset_next.png" border="0" /></html> | |
Repeat of Bradford for 10 nm Proteinase KRepeat Bradford analysis of Degradation of AuNP Fibers using 10 nm Proteinase K.
Bradford Analysis of Protease DegradationThe general procedure today was taken from Dr. Hartings's notebook. All samples will be incubated in eppendorf tubes, which will be placed in the 37°C water bath on the shaker. Samples should all contain the same concentration of materials. You should make samples for measurements at 10 minutes, 15 minutes, 30 minutes, 45 minutes, 1 hr, 1.5 hr, 2 hr. (Note, this is for the first time you do this. As you get better, I will expect you to get more measurements done).
DataThe spectrum for the overall blank with no protease or fiber sample was subtracted from all other spectra (blank and sample spectra). The spectra were further corrected by subtracting the absorbance at the isosbestic point. This data was redone from November 4th, because of the variation.
| |