User:Nkuldell/VitaYeast: Difference between revisions
| Line 68: | Line 68: | ||
#Prepare PCR cocktails. Each reaction will include: | #Prepare PCR cocktails. Each reaction will include: | ||
#*10 μl 2.5X PCR mastermix | #*10 μl 2.5X PCR mastermix | ||
#*0.5 μl | #*0.5 μl forward primer NO298 or NO300 | ||
#*0.5 μl | #*0.5 μl reverse primer NO299 or NO301 | ||
#*water to a final volume of 22.5 μl. | #*water to a final volume of 22.5 μl. | ||
#Add 22.5 μl of PCR cocktail to each PCR tube with yeast. | #Add 22.5 μl of PCR cocktail to each PCR tube with yeast. | ||
Revision as of 06:10, 10 June 2012
VitaYeast
From Boeke lab at Johns Hopkins, iGEM 2011 project
2007 paper describing Carotenoid production in S. cerevisiae is:
Appl Environ Microbiol. 2007 Jul;73(13):4342-50. Epub 2007 May 11.
High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous.
Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, van den Berg JA, van Ooyen AJ.
Source
Fungal Genomics, Laboratory of Microbiology, Wageningen University, Dreijenlaan 2, Wageningen, The Netherlands. pdf here
Initial communication about strains with Leslie Mitchell in Jef's lab
The authors supposedly sent us the four strains shown in Figure 4. I got a copy of these four strains from one of the iGEM-ers when I arrived in the lab - they were labelled 039, 128, 179, and 201. I can't figure out where these names came from and he wasn't too sure either, but I have continued to use the names. When the strains arrive in your lab I will use these labels as well.
In any case, I can give you the following information (this will make more sense once you read through the methods of the paper):
The four strains in Figure 4 should have the following genotypes:
- crtYB/I/E at URA3 - should be URA+ leu- trp-
- crtYB/I/E at URA3; Extra crtI at LEU2 - should be URA+ LEU+ trp-
- crtYB/I/E at URA3; tHMG1 at TRP - should be URA+ leu- TRP+
- crtYB/I/E at URA3; Extra crtI at LEU2; tHMG1 at TRP - should be URA+ LEU+ TRP+
in reality the 4 strains in our lab have the following genotypes:
- 179: URA+ LEU+ trp-
- 128: URA+ leu- TRP+
- 201: URA+ LEU+ TRP+
- 039: URA+ LEU+ TRP+
So clearly something doesn't quite jive between what is in the figure and the genotypes of the strains we have in the lab. But in any case, we do know for sure that all 4 strains do indeed contain at least one copy of crtYB/I/E - they all PCR amplify using primers specific for these genes. I've also attached a figure with the colours of the four strains compared to CEN.PK.
Cheers,
Leslie

Note: CEN.PK strain is described here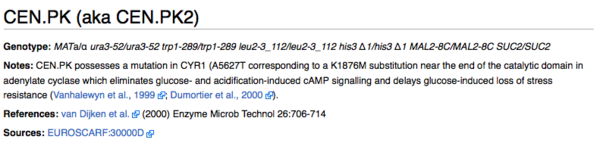
Getting started
- From Leslie:
- Filter paper method - I just cut up filter paper into little squares, wrap them individually in tin foil, then autoclave.
- When I want to send a strain, I resuspend a big scoop of cells in a small volume of media or water so I have a really dense 'culture', then spot ~50 uL onto the filter paper. re-wrap and send.
- To wake it a strain, I would dump the inoculated filter paper onto the appropriate plate, move it around a little with a sterile stick, and then streak for isolates using the stick. I usually repackage the filter paper in the tin foil just in case something goes wrong with the plate.
- Works like a charm for yeast and bacteria. I imagine either type of cell would be fine for several days, maybe a week, before streaking on media, but I have never done that experiment.
Summer 2012 Experiments
Primers
- Pr-crtYB-Tr-F
- NO298
- AAAACTGCAGCAGTTCGAGTTTATCATTATCAAT
- Pr-crtYB-Tr-R
- NO299
- TTTTCTGCAGGCGGCCGCGCAAATTAAAGCCTTCGAGCG
- Pr-crtI-Tr-F
- NO300
- CGGGGTACCCAGTTCGAGTTTATCATTATCAAT
- Pr-crtI-Tr-R
- NO301
- GCCGGTACCGGCCGGCCGCAAATTAAAGCCTTCGAGCG
Yeast Colony PCR
Protocols
Part 1: Colony PCR
From 20.109(F08) expt....
- Use the microwave to release the DNA from the yeast. On the tip of a sterile toothpick, pick-up 1/2 of the colony that is candidate and swirl the cells in 20 μl of sterile water in an eppendorf tube.
- Close the cap and microwave tube in an eppendorf rack for 15 seconds.
- Move 2.5 μl of the microwaved mixes, yeast debris and all, into a 200 μl PCR tube.
- Prepare PCR cocktails. Each reaction will include:
- 10 μl 2.5X PCR mastermix
- 0.5 μl forward primer NO298 or NO300
- 0.5 μl reverse primer NO299 or NO301
- water to a final volume of 22.5 μl.
- Add 22.5 μl of PCR cocktail to each PCR tube with yeast.
- Cycle the reactions as:
- 95° 4 minutes
- 95° 1 minute
- 52° 1 minute
- 72° 4 minutes
- repeat steps 2-4 35 times
- 72° 10 minutes
- 4° forever


