User:Nadine Rotundo/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| Line 17: | Line 17: | ||
http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/df903b12-a9dd-429d-bf46-bec83a5af713_zpsbvj2zvhp.jpg | http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/df903b12-a9dd-429d-bf46-bec83a5af713_zpsbvj2zvhp.jpg | ||
Image 1: | '''Image 1: ''' | ||
MB49 - NNNNNNNNNNNNNNNNANNNTGCAGCCGAGCGGTATTTGTCCTTCGGGACAGAGAGAGCGGCGTACGGGTGCGGAACACG TGTGCAACCTACCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATAAGTCAAGGCATCTTGAT TTATTGAAAACTCCGGTGGATAGAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCAATGA TCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGCAGTGAGG AATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGACGACGGCCCTATGGGTTGTAAACTTCTTTTG TATAGGGATAAACCTACTCTCGTGAGAGTANCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCAGCAGCCG CGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGGCTTGTAAGTCAGTGGTGA AATCTCATAGCTTAACTATGAAACTGCCATTGATACTGCAGGTCTTGAGTAAAGTAGAAGTGGCTGGAATAAGTAGTGTA GCGGTGAAATGCATAGATATTACTTANAACACCAATTGCGAAGGCAGGTCACTATGTTTTAACTGACGCTGATGGACGAA AGCGTGGGGAGCGAACAGGATTANATACCCTGGTAGTCCACGCCGTAAACGATGCTNACTCGTTTTTGGGCTTTCGGGTT CAGAGACTAAGCGAAAGTGATAAGTTAGCCACCTGGGGAGTACGTTCGCAAGAATGAAACTCAAAGGAATTGACGGGGGC CCGCACAANCGGTNNTTATGTGGNTTAATTCGATGATANNCGAGGAACCTTANCAAAGGCTNAAATGGGAATTGACAGGN TTANAAAATAGACTTTTCTTCNNACNATTTTCAAGNTGCTGCATGGNNGTCNNCAGCTCGTGCCNTGAGTGTNGNTAAGT CCTGCAACNANCNCAACCCNGNNNNTANNTNNCATNNTTCAGTTNGGGANNNNTAGNNNN | MB49 - NNNNNNNNNNNNNNNNANNNTGCAGCCGAGCGGTATTTGTCCTTCGGGACAGAGAGAGCGGCGTACGGGTGCGGAACACG TGTGCAACCTACCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATAAGTCAAGGCATCTTGAT TTATTGAAAACTCCGGTGGATAGAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCAATGA TCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGCAGTGAGG AATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGACGACGGCCCTATGGGTTGTAAACTTCTTTTG TATAGGGATAAACCTACTCTCGTGAGAGTANCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCAGCAGCCG CGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGGCTTGTAAGTCAGTGGTGA AATCTCATAGCTTAACTATGAAACTGCCATTGATACTGCAGGTCTTGAGTAAAGTAGAAGTGGCTGGAATAAGTAGTGTA GCGGTGAAATGCATAGATATTACTTANAACACCAATTGCGAAGGCAGGTCACTATGTTTTAACTGACGCTGATGGACGAA AGCGTGGGGAGCGAACAGGATTANATACCCTGGTAGTCCACGCCGTAAACGATGCTNACTCGTTTTTGGGCTTTCGGGTT CAGAGACTAAGCGAAAGTGATAAGTTAGCCACCTGGGGAGTACGTTCGCAAGAATGAAACTCAAAGGAATTGACGGGGGC CCGCACAANCGGTNNTTATGTGGNTTAATTCGATGATANNCGAGGAACCTTANCAAAGGCTNAAATGGGAATTGACAGGN TTANAAAATAGACTTTTCTTCNNACNATTTTCAAGNTGCTGCATGGNNGTCNNCAGCTCGTGCCNTGAGTGTNGNTAAGT CCTGCAACNANCNCAACCCNGNNNNTANNTNNCATNNTTCAGTTNGGGANNNNTAGNNNN | ||
Revision as of 22:23, 2 March 2015
PCR - March 5th, 2015
PURPOSE: The purpose of the PCR for 16S sequencing was to understand how DNA sequences are used to identify species.
MATERIALS & METHODS:
Procedure 1: In a previous lab, we plated four separate agar plates. Two out of the four plates contained the tetracycline antibiotic. We characterized bacteria from two of the plates and used primers and PCR to amplify the 16S rRNA gene. This helped us identify the types of bacteria that were present in our transect.
Procedure 2: From two out of the four plates, we transferred a single colony of bacteria to a 100 ul of water in a sterile tube. We then incubated the tubes at 100ºC for 10 minutes in a heat block. We then centrifuged the samples for 5 minutes at 13,400 rpm. During the centrifuge, we added 20 ul of primer/water mixture to a labeled PCR tube and mixed the sample until the bead was dissolved. We then transferred 5 ul of supernatant from our centrifuged samples to the 16S PCR reaction and placed the sample into the PCR machine.
Procedure 3: The following week, we ran the PCR product on an agarose gel. Since we had a PCR product form, our professor purified the DNA for sequencing.
DATA AND OBSERVATIONS
Image 1:
MB49 - NNNNNNNNNNNNNNNNANNNTGCAGCCGAGCGGTATTTGTCCTTCGGGACAGAGAGAGCGGCGTACGGGTGCGGAACACG TGTGCAACCTACCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATAAGTCAAGGCATCTTGAT TTATTGAAAACTCCGGTGGATAGAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCAATGA TCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGCAGTGAGG AATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGACGACGGCCCTATGGGTTGTAAACTTCTTTTG TATAGGGATAAACCTACTCTCGTGAGAGTANCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCAGCAGCCG CGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGGCTTGTAAGTCAGTGGTGA AATCTCATAGCTTAACTATGAAACTGCCATTGATACTGCAGGTCTTGAGTAAAGTAGAAGTGGCTGGAATAAGTAGTGTA GCGGTGAAATGCATAGATATTACTTANAACACCAATTGCGAAGGCAGGTCACTATGTTTTAACTGACGCTGATGGACGAA AGCGTGGGGAGCGAACAGGATTANATACCCTGGTAGTCCACGCCGTAAACGATGCTNACTCGTTTTTGGGCTTTCGGGTT CAGAGACTAAGCGAAAGTGATAAGTTAGCCACCTGGGGAGTACGTTCGCAAGAATGAAACTCAAAGGAATTGACGGGGGC CCGCACAANCGGTNNTTATGTGGNTTAATTCGATGATANNCGAGGAACCTTANCAAAGGCTNAAATGGGAATTGACAGGN TTANAAAATAGACTTTTCTTCNNACNATTTTCAAGNTGCTGCATGGNNGTCNNCAGCTCGTGCCNTGAGTGTNGNTAAGT CCTGCAACNANCNCAACCCNGNNNNTANNTNNCATNNTTCAGTTNGGGANNNNTAGNNNN Chryseobacterium sp. LDVH 3 16S ribosomal RNA gene, partial sequence
MB50 - NNNNNNNNNNNNNNNNNNNNNANNNNTGCAGCCGAGCGGTATTGTTTCTTCGGAAATGAGAGAGCGGCGTACGGGTGCGG ANCNNNTGTGCAACCTGCCTTTATCTGGGGGATAGCCTTTCGAAAGGGAGATTAATACCCCATAATATATTAAGTGGCAT CACTTGATATTGAAAACTCCGGTGGATAGAGATGGGCACGCGCAAGATTAGATAGTTGGTGAGGTAACGGCTCACCAAGT CTACGATCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGCA GTGAGGAATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGACGACGGCCCTATGGGTTGTAAACTT CTTTTGTATAGGGATAAACCTACTCTCGTGAGAGTAGCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCAG CAGCCGCGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAAGGGTCCGTANGCGGATCTGTAAGTCA GTGGTGAAATCTCACAGCTTAACTGTGAAAACTGCCATTGATACTGCAGGTCTTGAGTGTTGTTGAAGTANCTGGAATAA GTAGTGTANCGGTGAAATGGCNTAGATATTACTTAGAAACACCAATTGCNAAGGCTNGTTACTAANCAACAACTGACNCT GATGGACGAAANCGTGGNGGAGCGAACAGGATTANATACCCCTGGNAN Chryseobacterium sp. StRB028 gene for 16S rRNA, partial sequence
CONCLUSIONS & FUTURE DIRECTIONS
The two bacteria that had the closest match after DNA sequencing were the bacteria listed above. Since some of the bacteria show tetracycline resistance, it suggests that our transect has been exposed to antibiotics previously. This could have derived from runoff of from pesticides or other chemicals directly applied to the area.
Embryology & Zebrafish Development - February 26, 2015
PURPOSE: The purpose of this lab was to study the stages of embryonic development of zebrafish and compare it with the development in different organisms. We also set up a baseline experiment to help us study how exposure to Ret A affects embryonic development in zebrafish.
MATERIALS & METHODS:
After reading a published study that went into great detail about the adverse effects that different amounts of Ret A exposure has on the embryonic development of zebrafish, we are ready to set up our own experiment.
PROCEDURE 1: We observed and analyzed the embryonic development of starfish, frogs, and chicks under a dissecting microscope. We compared the embryological features including egg size, type of fertilization, blastulation, gastrulation, and more.
PROCEDURE 2: We then created a control and an experimental group in covered petri dishes. For the control, we added 20 mLs of distilled water; for the experimental group, we added 10mL of Ret A and 10mL of distilled water. We then added 20 healthy embryos were added to each dish. These embryos were at the stage of 40% of development. The embryos are to be observed at 4, 7, and 11 days following the procedure to observe the effects of Ret A at different points of development.
DATA AND OBSERVATIONS
Image 1: Day 1 Control Petri Dish (left) and Experimental Ret A Petri Dish (right)
http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/Biology210_RetA_Day1.jpg_zpszdnttl7a.png
Image 2: Day 4 Control Fish (40x)
Image 3: Day 4 Ret A Fish (40x)
Image 4: Day 4 Ret A Fishes
http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/800px-Biology210_RetA_Day4_zpsx7lilaw2.jpeg
CONCLUSIONS & FUTURE DIRECTIONS
At day 4 of the Ret A exposure, there are only slightly noticeable changes in the body structure of the fish: the Ret A fish are slightly more plump. While these are only the changes in visible development, we predicted that if the zebrafish embryos are exposed to an excess concentration of Ret A then they would also exhibit heart, brain, and embryonic deformities as well. We hypothesized that the relationship between Ret A and body function has a dose-response relationship, and the higher the exposure of the drug, the more deformities the zebrafish embryos the will possess.
2.20.15 Excellent entry. Clear results. Good food web. SK
Lab 5 - Invertebrates
- February 19
PURPOSE: To understand the importance of invertebrates and learn how their systems evolved into more complex systems over time.
MATERIALS & METHODS:
Procedure 1: We first observed acoelomates, pseudocoelomates, and coelomates with a dissecting scope to observe how they digested an egg yolk.
While all worms fall under the broad phylum of anthropods, there are three distinct types of worms. Body structure relates directly to function and each worm operates in different ways specific to their shape, size, and muscle and sensory anatomy. The strength of the sensory and digestive system and the relationship between them is vital in how an organism moves and survives. Muscle and sensory system coordination is needed for an organism to get food, avoid predators, and survive in a given environment or transect.
Acoelomates have a very simple digestive system; they lack a coelom, a fluid filled cavity that separates the digestive tract from the inside wall of their digestive system. Acoelomates also lack the structural complexity needed to form a fully functioning sensory system. Pseudocoelomates have an incompletely lined body cavity, coelom, which allows for the organisms to have a very basic circulatory system. Coelomates have a fully lined and fully filled body cavity, which allows them to have a fully functioning sensory and digestive system. Acoelomates, Pseudocoelomates, and Coelomates are in ascending order of complexity, which respectively demonstrates their ability to maneuver through soil.
Procedure 2:
We then observed organisms from each of the five major classes and observed differences in body parts, body segments, and the number of appendages the organisms had.
Procedure 3:
Then we observed the invertebrates that were collected from our Berlese Funnel and found in our transect. We examined the organisms by eye and with a microscope, and used a key to help us identify what was in our petri dish.
Most soil inverterbrates are in the phylum anthropod, which include a wide range of species. We can expect to find microanthropods, which are mites or pseudoscorpians and are closely related to spiders. We can also expect to find worms, either Nematodes (roundworms) or earthworms. Lager anthropods including centipedes, beetles, spiders, ants, and other species often commonly live in soil. Most anthropods are considered predators by nature because they survive off the resources that the transects provide. They feast off roots, fungi, and decaying plant material, which is destructive to the viability of the transect, but also allows for evolutionary mechanisms to take place.
Procedure 4:
By definition, vertebrates are a classification of animals that have a spinal cord that contains nerve cells that run from the brain to the posterior of the body. Some vertebrates that have been seen in our transect or are expected to inhabit it are humans, small mammals, large mammals, and birds. While we have only observed in person squirrels and a Blue Jay in our transect, I used to live in Leonard Hall, which overlooks our transect, and I have heard and seen Woodpeckers, rats, and a raccoon first hand.
DATA AND OBSERVATIONS:
By using the Berlese Funnel, we were able to collect 5 invertebrates from ourtransect. Our collection of invertebrates consisted of 5 insects and 3 separate species demonstrated in the table below.
Table 1: Soil Inverterbrates found in Transect 3
The organisms we observed ranged from 1/10th of a centimeter to 17/10th of a centimeter. All organisms were able to be seen with a naked eye. The largest organism that we found in our transect was a centipede, mainly because it has the longest body out of all the common anthropods. The smallest were the ground spiders, which were very hard to find because of their small size and light color.
Image 1: Ground Spider
Image 2: 2 Springtails
Image 3: Centipede
Table 2: Classification of Species found in our transect
Table 3: Classification of biotic and abiotic characteristics beneficial to species
Figure 1: Food Web for Organisms located in our transect.
These organisms accurately represent the dynamics of a miniature ecosystem. The definition of an ecological community is an environment shared by two or more different species. As depicted in the image above, there is a variety of different microorganisms, insects, and animals that all share the transect. A carrying capacity is the maximum number of species and organisms that can occupy a space successfully without running out of water and resources. When the number of species present exceed the carry capacity, competition for resources begin and the cycle of natural selection and evolution continues. Every ecosystem contains a pattern of trophic levels that basically describes the food chain that depicts how organisms survive in an ecosystem. The food web above demonstrates which organisms are preys and predators, and how organisms obtain the resources needed to survive. A chart of the trophic levels of our transect can be shown below.
Table 4: Trophic levels of organisms found in our transect.
CONCLUSIONS:
I think the purpose of this lab was to demonstrate the many different factors that are present in an ecosystem that allows it to function and survive. A wide variety of both microorganisms and microorganisms, ranging from bacteria, fungi, protists, invertebrate, and vertebrate, and more survive off the available resources and both biotic and abiotic features that our 20x20 area provides them. It is both amazing and fascinating that so much biological activity takes place in our transect without us even knowing it.
2.20.15 Good entry. It would be improved by including the data table from your manual with more information on your plants of interest. SK
Lab 4 - Plantae and Fungi - - February 12:"
PURPOSE: The purpose of the current lab is to understand the characteristics and diversity of Plants
MATERIALS & METHODS: In order to accurately identify and study the plants that live inside our transect, we had to carry out a multistep procedure. The procedure included 1) observing characteristics of a Bryophute moss, Mnium; 2) an angiosperm, Lilium; and 3) five plant samples from our transect. We categorized all of these samples off of three general features: the presence of vascularization, the presence of Specialized structures, and the mechanism of reproduction.
Procedure I: First, we obtained a random leaf and soil sample from our transect. We then derived five separate types of plants from this matter to use for observations. The rest was saved for our Berlese funnel later on in the experiment.
Procedure II: Secondly, we compared the heights of the moss, Mnium, and the lily plant stem.
Procedure III: Next, we examined the leaves of the moss using a dissection scope.
Procedure IV: Then, we compared the reproductive cycles of the plants, paying specific attention to whether the plant was haploid or diploid.
Procedure V: Then we observed fungi, specifically Rhizopus Stolonifer, a common zygomycetes. Some fungi contain small, black, circular structures called sporangia. These sporangia are made up of smaller cells called spores. Sporangia can be found on common types of mold, such as zygomycetes, which is infamously known as the black bread mold. It is referred to as the black bread mold because the sporangia turn black upon maturation. These sporangia and mold can be seen in the image below.
Image 6: Rhizopus Stolonifer, a common zygomycetes, also known as black bread mold.
Procedure VI: Lastly, we set up the Berlese Funnel to collect the invertebrates in our sample. We poured 25 mL of the 50:50 ethanol/water solution into the 50 mL conical tube. Then we fit a piece of the screening material into the bottom of the funnel. We added our leaf matter into the top of the funnel and set it securely into the ring stand. An ethanol tube was attached to the bottom of the funnel so the ethanol will not evaporate. Then a 40 watt lamp was placed above the funnel and the entire structure was covered with foil. We left the Berlese Funnel on the lab bench for one week and will revisit it during our next lab session. The final structure can be viewed below.
DATA & OBSERVATIONS:
[[Image:[URL=http://s1379.photobucket.com/user/Nadine_Rotundo/media/10978621_10204583346157117_3996643702295370363_n_zpsugnugjhx.jpg.html][IMG]http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/10978621_10204583346157117_3996643702295370363_n_zpsugnugjhx.jpg[/IMG][/URL]]]
Image 1: Plant #1: Located on shrub, approximately 500 found in our transect. Its size is approximately 5x3.5 mm. The plant is red, green, and orange. It appears to be during and the sides are curling. The vascularization of this plant is monocot, it has no specialized structures, and its mechanism of reproduction is alteration of genetics.
[[Image:[URL=http://s1379.photobucket.com/user/Nadine_Rotundo/media/28283b51-c1c0-4dde-872b-af366abe4fb1_zpsov0wlowk.jpg.html][IMG]http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/28283b51-c1c0-4dde-872b-af366abe4fb1_zpsov0wlowk.jpg[/IMG][/URL]]]
Image 2: Plant #2: Located on the ground, approximately 500 found in our transact. Approximately 6x2.5 mm in size. The leaf is green spotted with brown spots; it appears to be dying and has wrinkled edges. The vascularization of this plant is monocot, it has no specialized structures, and its mechanism of reproduction is alteration of genetics.
[[Image:[URL=http://s1379.photobucket.com/user/Nadine_Rotundo/media/10387208_10204583346437124_2035102533959822300_n_zps2nlupz6x.jpg.html][IMG]http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/10387208_10204583346437124_2035102533959822300_n_zps2nlupz6x.jpg[/IMG][/URL]
]]
Image 3: Plant #3: Located on the ground, approximately 500 found in our transect. Approximately 4x5.5 mm in size. The leaf is grayish-brown, appears to be dying, and is cracking at the edges. The vascularization of this plant is dicot, it has no specialized structures, and its mechanism of reproduction is alteration of genetics.
[[Image:[URL=http://s1379.photobucket.com/user/Nadine_Rotundo/media/10950699_10204583346717131_154076383926522631_n_zpsy5r8vl0o.jpg.html][IMG]http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/10950699_10204583346717131_154076383926522631_n_zpsy5r8vl0o.jpg[/IMG][/URL]]]
Image 4: Plant #4: Located on the ground, approximately 300 found in the transect (slightly less prominent than the previous plants). Its size is approximately 4.5 x 2 mm. The leaf is pours, brown, dead, and brittle, but it does have strong structural definition. The vascularization of this plant is monocot, the only specialized strutters that it has is a net with defined edges, and its mechanism of reproduction is alteration of genetics.
Image 5: Plant #5: Located on a tree. Approximately 75 were found in the transect (not very many at all). Its size is 4x1.5 (per individual leaf), and all leaves were very fresh, green colored, and seemed alive. The texture of the leaves were intact and smooth. The vascularization of this plant is monocot, it has no specialized structures, and its mechanism of reproduction is alteration of genetics.
[[Image:[URL=http://s1379.photobucket.com/user/Nadine_Rotundo/media/10612791_10204583347077140_4723469001341755998_n_zps53urle5s.jpg.html][IMG]http://i1379.photobucket.com/albums/ah152/Nadine_Rotundo/10612791_10204583347077140_4723469001341755998_n_zps53urle5s.jpg[/IMG][/URL]
]]
CONCLUSIONS & FUTURE DIRECTIONS:
This procedure allowed us to see the wide range of microorganisms and microorganisms that are living in our transect. In our next lab, we will be able to further this investigation by testing which vertebrates are inhabiting the land that we are observing.
2.10.15 Good entry but missing some detail. Need to include description of colony morphology and 16s PCR set up. Data tables would allow you present your results clearly. Try using http://excel2wiki.net to convert excel tables into tables for OWW. SK
Lab 3 - February 5
== Purpose: ==
The purpose of this lab was to understand the characteristics of bacteria and observe antibiotic resistance to microorganisms found in our transect.
Methods:
In our previous lab, we prepped two sets of agar plates. The first set only contained nutrients with four different levels of dilution; the second set was the same as this but also included the tetracycline antibiotic. After a week, colonies of bacteria were visible on our agar plates. We then analyzed these colonies using the bacteria morphology key to help identify what we were looking at. See the image below.
We then took samples of the bacteria we observed on the agar plate to test whether they were gram negative or gram positive bacteria by using the Gram Stain Procedure. The first step of the Gram Stain Procedure is to sterilize the hook by moving it over an open flame, then picking up the bacteria with the loop. The bacteria is then smeared onto a slide and dried over the flame until it is securely dried to the slide. Then, over a working tray, the bacteria smear is covered with crystal violet for 1 minute, then rinsed with water; then covered with Gram's iodine for 1 minute, then rises with water again; then rinsed with 95% alcohol for 10-20 seconds; then the slide is covered with safranin stain for 20-30 seconds, then rinsed. The excess liquid on the slide was blotted and then it was ready to be observed underneath the microscope.
==
Data ==
Image 1: The agar plates containing the TET is located on the lower level of the image. Notice that there are not as many bacteria colonies present.
Image 2: This is a closeup on the two agar plates that we decided to look into further detail for. Both are concentrations of 10^-5.
'Table 1:' 100-fold Serial Dilutions Results- A written summary of what we observed on our agar plates.
Table 2: Bacteria Characterization - A summary of the bacteria that we observed and analyzed.
==
Conclusions ==
The Gram Stain Procedure allowed us to determine which bacteria were gram positive and which were gram negative. Gram negative bacteria show up under the microscope as pink, and gram positive bacteria show up under the microscope as purple. Since both of the TET plates contained purple microorganisms, we were able to derive that gram positive bacteria are antibiotic resistant. The slides with no antibiotic and just nutrient both contained pink, gram negative organisms.
The last part of our experiment was to set up a PCR procedure for this week's lab. The results to this procedure will help us understand how DNA sequences are used to identify the species in our slides.
2.4.15 Good start. The notebook should be organized into: Purpose, Methods, Data & conclusions using headings. The pictures are too big. Try using smaller image files to upload. Could include more detail on specific protists identified from Hay Infusion, where in the Hay Infusion they were obtained from and more descriptionSK
January 29 - Identifying Algae and Protists
Our culture had floating white stuff in it, which is expected to be mold. The sample smelled very strongly and badly and the water was murky. There was distinct differences between the top, middle, and bottom section of the hay infusion sample. The top had a slim white film cover, the middle was cloudy green with some leaves and grass floating, and the bottom was densely populated with leaves, muck, and other algae. In the top portion of our sample we found chlamydomonas and euplotes sp. In the middle portion we found Bursaria truncatella and paramecium aurelia. In the bottom sample we found paramecium Bursaria and vorticella. I have included sketches of both the paramecium burs aria and bursaria truncatell below.
Every organism needs specific conditions to be able to survive. The main components that are needed are food, water, oxygen, living space, and the ability to reproduce. Paramecium do not perform photosynthesis, have two nuclei, and can reproduce sexually or asexually.
If the Hay Infusion Culture grew for another two months then it would eventually run out of sources of nutrients and die off because it is harder for microorganisms to survive in an isolated community.
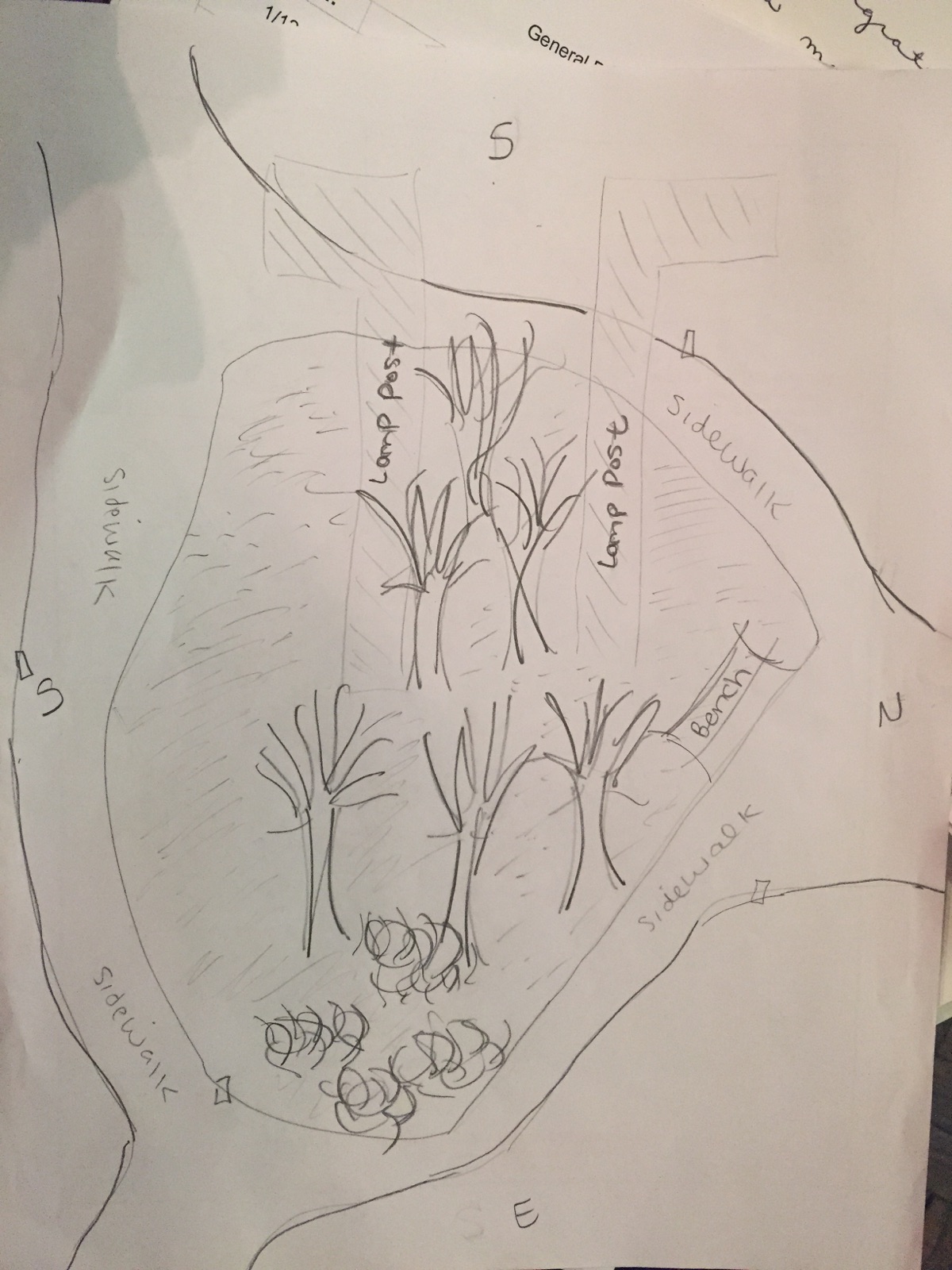
January 22, 2015 - Biological Life at AU
Our transect is located in the center of campus in the amphitheater woods. The location is densely populated with trees and other microorganisms given the nature of American University's campus in general. There are also some built components to the transect, including a paved sidewalk, various benches, and some lamp posts. The transect is also often visited by squirrels, rats, raccoons (I previously lived in Leonard Hall near here and can speak for this), insects, and college students.
As I touched on previously, the biotic features in our transect include leaves, trees, bushes and brush, squirrels, rats, raccoons, and humans. The abiotic components of our transect include metal lamp posts, a concrete sidewalk, stones, cigarette butts, and a discarded ribbon (litter). As indicated in the image, the leaves, trees, brush and bushes are found in the center of the transect, which is overall surrounded by the concrete sidewalk. All the animals I mentioned earlier visit the center of the transect, but humans are assumed to remain on the sidewalk and benches. The lamp posts are located directly in the center of the transect as well, surrounded by many trees. The transect has a few inclines and levels.