User:Musfeq Us Salehin Patoary/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
Data and Observations: | Data and Observations: | ||
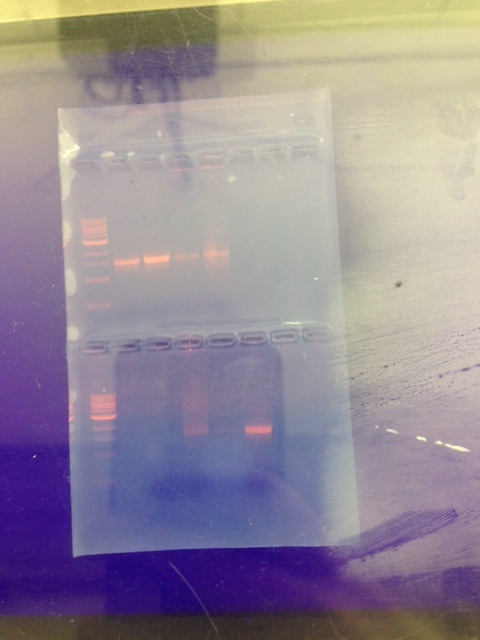
[[Image:16s.jpg]] | |||
Sequence for MB59 NNNNNNNNNNNNNNNNNNAGCNNNGCAGTCGAGCGGANGANGGGAGCTTGCTCCTTGATTCAGCGGCGGACGGGTGAGTA ATGCCTAGGAATCTGCCTGGTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGTCCTACGGGAGAAAGC AGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCGGATTAGCTAGTTGGTGGGGTAATGGCTCACCAAGGCGA CGATCCGTAACTGGTCTGAGAGGATGATCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTG GGGAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGATTGTAAAGCACTTT AAGTTGGGAGGAAGGGCAGTAAGCTAATACCTTGCTGTTTTGACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCC ANCANCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTANNTGGTTTGTTNNGTT NNATGTGAAANCCCCNGGGCTCAACCTGNGGAACTGCATCCCNNACTGGNNNNCTANANTACGGTGNANGGTGNTGNAAT TTCNTGTGNNGNNNTGAAATGCNTAGATANANCATTAACAGCGGNGACNANGGGACNNCNTNNACTNGTTNGNNCCNCNN ATGTGCGNNNNCNGGTGNNGNAAACANNNNNGAGNNCCNCCTTCGTCNNNGNNNNNNNNANNATNNNNCTACCCCNTTNN NNNGNTNNNNGNANTTNCCNNGCCTNNCCNCNNGANNNNANCNNNNCGTTNNNNNAGCNCNCCNTNGNGNNNACNGGNNT TTNNNAGNCNNNGNNNANNCAACNNNNNNAGNNNNNGTNGNNCNANTNNNNNNAAAANNNATNNNNNNNNCCNTNNNNTA GNACGNNNGACGNNNNNNNNANNNNGNNNNN | Sequence for MB59 NNNNNNNNNNNNNNNNNNAGCNNNGCAGTCGAGCGGANGANGGGAGCTTGCTCCTTGATTCAGCGGCGGACGGGTGAGTA ATGCCTAGGAATCTGCCTGGTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGTCCTACGGGAGAAAGC AGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCGGATTAGCTAGTTGGTGGGGTAATGGCTCACCAAGGCGA CGATCCGTAACTGGTCTGAGAGGATGATCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTG GGGAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGATTGTAAAGCACTTT AAGTTGGGAGGAAGGGCAGTAAGCTAATACCTTGCTGTTTTGACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCC ANCANCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTANNTGGTTTGTTNNGTT NNATGTGAAANCCCCNGGGCTCAACCTGNGGAACTGCATCCCNNACTGGNNNNCTANANTACGGTGNANGGTGNTGNAAT TTCNTGTGNNGNNNTGAAATGCNTAGATANANCATTAACAGCGGNGACNANGGGACNNCNTNNACTNGTTNGNNCCNCNN ATGTGCGNNNNCNGGTGNNGNAAACANNNNNGAGNNCCNCCTTCGTCNNNGNNNNNNNNANNATNNNNCTACCCCNTTNN NNNGNTNNNNGNANTTNCCNNGCCTNNCCNCNNGANNNNANCNNNNCGTTNNNNNAGCNCNCCNTNGNGNNNACNGGNNT TTNNNAGNCNNNGNNNANNCAACNNNNNNAGNNNNNGTNGNNCNANTNNNNNNAAAANNNATNNNNNNNNCCNTNNNNTA GNACGNNNGACGNNNNNNNNANNNNGNNNNN | ||
Sequence for MB60 NNNNNNNNNNNGNNNGCTTNNNNTGCAGTCGAGCGGGGAGATGTAGCTTGCTACATTTCCTAGCGGCGGACGGGTGAGTA ATGCTTAGGAATCTGCCTATTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGCCCTACGGGGGAAAGC AGGGGATCTTCGGACCTTGCGCTAATAGATGAGCCTAAGTCAGATTAGCTAGTTGGTGGGGTAAAGGCCTACCAAGGCGA CGATCTGTAGCGGGTCTGAGAGGATGATCCGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTG GGGAATATTGGACAATGGGGGGAACCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGCCTTTTGGTTGTAAAGCACTTT AAGCGAGGAGGAGGCTACTAGTACTAATACTACTGGATAGTGGACGTTACTCGCAGAATAAGCACCGGCTAACTCTGTGC CAGCAGCCGCGGTAATACAGAGGGTGCGAGCGTTAATCGGATTTACTGGGCGTAAAGCGTACGTAGGCGGCTTTTTAAGT CGGATGTGAAATCCCTGAGCTTAACTTAGGANTTGCATTCGATACTGGGAAGCTAGAGTATGGGAGAGGATGGTAGAATT CCAGGTGTAGCGGTGAAATGCGTAGAGATCTGGAGGAATACCNATGGCNAANGCGGCCATCTGGCCTGNTACTGACGCTN ANGTACGAAANCATGGGGAGCANACAGGATTAGATACCCTGNTAGTCCATGCCNTANNCNATGTCTACTNNCCNTTGGGG CCNNTGANNNNNTANTNNCNCAGCTCACGCNATAANTNNACCGCCTGGGNAGTGNNGNNCNCAGNNTANACTCAAATGNN TTGACANNGNCCNGCACAANCGNTNGANCATGNGNNTTTNNNTCNATGCANNNNNANNNCCNTNACCTGCTNNTTGNNGN TANNNNNNANCNNNNNNGGNANATNNNNTNGGNGGCGNTNGNTNANCTTNNNGNANANNTNNCNNCNTNGNNNNGNANGN GCTNNNNGTCNNCNNNTGNNNGNNNANNNNNGGNNNAANNNNNNNNNNNNNNNNNNTNNNNNTNNNNNN | |||
Sequence for MB60 NNNNNNNNNNNGNNNGCTTNNNNTGCAGTCGAGCGGGGAGATGTAGCTTGCTACATTTCCTAGCGGCGGACGGGTGAGTA ATGCTTAGGAATCTGCCTATTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGCCCTACGGGGGAAAGC AGGGGATCTTCGGACCTTGCGCTAATAGATGAGCCTAAGTCAGATTAGCTAGTTGGTGGGGTAAAGGCCTACCAAGGCGA CGATCTGTAGCGGGTCTGAGAGGATGATCCGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTG GGGAATATTGGACAATGGGGGGAACCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGCCTTTTGGTTGTAAAGCACTTT AAGCGAGGAGGAGGCTACTAGTACTAATACTACTGGATAGTGGACGTTACTCGCAGAATAAGCACCGGCTAACTCTGTGC CAGCAGCCGCGGTAATACAGAGGGTGCGAGCGTTAATCGGATTTACTGGGCGTAAAGCGTACGTAGGCGGCTTTTTAAGT CGGATGTGAAATCCCTGAGCTTAACTTAGGANTTGCATTCGATACTGGGAAGCTAGAGTATGGGAGAGGATGGTAGAATT CCAGGTGTAGCGGTGAAATGCGTAGAGATCTGGAGGAATACCNATGGCNAANGCGGCCATCTGGCCTGNTACTGACGCTN ANGTACGAAANCATGGGGAGCANACAGGATTAGATACCCTGNTAGTCCATGCCNTANNCNATGTCTACTNNCCNTTGGGG CCNNTGANNNNNTANTNNCNCAGCTCACGCNATAANTNNACCGCCTGGGNAGTGNNGNNCNCAGNNTANACTCAAATGNN TTGACANNGNCCNGCACAANCGNTNGANCATGNGNNTTTNNNTCNATGCANNNNNANNNCCNTNACCTGCTNNTTGNNGN TANNNNNNANCNNNNNNGGNANATNNNNTNGGNGGCGNTNGNTNANCTTNNNGNANANNTNNCNNCNTNGNNNNGNANGN GCTNNNNGTCNNCNNNTGNNNGNNNANNNNNGGNNNAANNNNNNNNNNNNNNNNNNTNNNNNTNNNNNN | |||
MB59 Species- Pseudomonas putida strain YN3 | MB59 Species- Pseudomonas putida strain YN3 | ||
MB60 Species- Acinetobacter bouvetii strain CCM7196 | MB60 Species- Acinetobacter bouvetii strain CCM7196 | ||
Conclusions: | |||
Pseudomonas putida and Actinobactor bouvetti were sequenced. The Pseudomonas putida is a gram negative, rod shaped probacterium . The Actinobactor genus is gram negative,nonmotile,non fermenting cocci or coccobacilli. | |||
MP | |||
Revision as of 14:12, 5 March 2015
March 5th,2015 16s DNA sequencing
Purpose: The purpose is to determine the species of bacteria in transect 5.
Materials and Methods: A PCR reaction for the 16s gene was run on our bacterial samples collected from transect 5.Two samples from nutrient agar plate were sent off for sequencing and the resulting sequences were used in NCBI Blast program to determine the species of bacteria.
Data and Observations:
Sequence for MB59 NNNNNNNNNNNNNNNNNNAGCNNNGCAGTCGAGCGGANGANGGGAGCTTGCTCCTTGATTCAGCGGCGGACGGGTGAGTA ATGCCTAGGAATCTGCCTGGTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGTCCTACGGGAGAAAGC AGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCGGATTAGCTAGTTGGTGGGGTAATGGCTCACCAAGGCGA CGATCCGTAACTGGTCTGAGAGGATGATCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTG GGGAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGATTGTAAAGCACTTT AAGTTGGGAGGAAGGGCAGTAAGCTAATACCTTGCTGTTTTGACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCC ANCANCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTANNTGGTTTGTTNNGTT NNATGTGAAANCCCCNGGGCTCAACCTGNGGAACTGCATCCCNNACTGGNNNNCTANANTACGGTGNANGGTGNTGNAAT TTCNTGTGNNGNNNTGAAATGCNTAGATANANCATTAACAGCGGNGACNANGGGACNNCNTNNACTNGTTNGNNCCNCNN ATGTGCGNNNNCNGGTGNNGNAAACANNNNNGAGNNCCNCCTTCGTCNNNGNNNNNNNNANNATNNNNCTACCCCNTTNN NNNGNTNNNNGNANTTNCCNNGCCTNNCCNCNNGANNNNANCNNNNCGTTNNNNNAGCNCNCCNTNGNGNNNACNGGNNT TTNNNAGNCNNNGNNNANNCAACNNNNNNAGNNNNNGTNGNNCNANTNNNNNNAAAANNNATNNNNNNNNCCNTNNNNTA GNACGNNNGACGNNNNNNNNANNNNGNNNNN
Sequence for MB60 NNNNNNNNNNNGNNNGCTTNNNNTGCAGTCGAGCGGGGAGATGTAGCTTGCTACATTTCCTAGCGGCGGACGGGTGAGTA ATGCTTAGGAATCTGCCTATTAGTGGGGGACAACGTTTCGAAAGGAACGCTAATACCGCATACGCCCTACGGGGGAAAGC AGGGGATCTTCGGACCTTGCGCTAATAGATGAGCCTAAGTCAGATTAGCTAGTTGGTGGGGTAAAGGCCTACCAAGGCGA CGATCTGTAGCGGGTCTGAGAGGATGATCCGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTG GGGAATATTGGACAATGGGGGGAACCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGCCTTTTGGTTGTAAAGCACTTT AAGCGAGGAGGAGGCTACTAGTACTAATACTACTGGATAGTGGACGTTACTCGCAGAATAAGCACCGGCTAACTCTGTGC CAGCAGCCGCGGTAATACAGAGGGTGCGAGCGTTAATCGGATTTACTGGGCGTAAAGCGTACGTAGGCGGCTTTTTAAGT CGGATGTGAAATCCCTGAGCTTAACTTAGGANTTGCATTCGATACTGGGAAGCTAGAGTATGGGAGAGGATGGTAGAATT CCAGGTGTAGCGGTGAAATGCGTAGAGATCTGGAGGAATACCNATGGCNAANGCGGCCATCTGGCCTGNTACTGACGCTN ANGTACGAAANCATGGGGAGCANACAGGATTAGATACCCTGNTAGTCCATGCCNTANNCNATGTCTACTNNCCNTTGGGG CCNNTGANNNNNTANTNNCNCAGCTCACGCNATAANTNNACCGCCTGGGNAGTGNNGNNCNCAGNNTANACTCAAATGNN TTGACANNGNCCNGCACAANCGNTNGANCATGNGNNTTTNNNTCNATGCANNNNNANNNCCNTNACCTGCTNNTTGNNGN TANNNNNNANCNNNNNNGGNANATNNNNTNGGNGGCGNTNGNTNANCTTNNNGNANANNTNNCNNCNTNGNNNNGNANGN GCTNNNNGTCNNCNNNTGNNNGNNNANNNNNGGNNNAANNNNNNNNNNNNNNNNNNTNNNNNTNNNNNN
MB59 Species- Pseudomonas putida strain YN3
MB60 Species- Acinetobacter bouvetii strain CCM7196
Conclusions:
Pseudomonas putida and Actinobactor bouvetti were sequenced. The Pseudomonas putida is a gram negative, rod shaped probacterium . The Actinobactor genus is gram negative,nonmotile,non fermenting cocci or coccobacilli.
MP
February 18th, 2015- Vertebrates
Purpose: We are in group studding a small area (transect 5) of American University to understand species. In this lab, we are focusing on vertebrates.
Materials and Methods: It is winter season and weather is very cold and less plants and insects in our transect 5 compare to summer. There are no vertebrate in our transect five at the time we were there but we found some species around that transect . Those vertebrates moving constantly for foods. I believe they sometimes search foods in transect 5 also. We observe them and studied at home to find the classifications.
Data and observations: I observed five vertebrates species as follows:
Bald Eagle (genus Haliaeetus, Order Accipitriformes, Haliaeetus Leuciceorialis) Resting on a top of a tree
Eastern Fox Squirrel (Phylum Cordata, Sciurus Niger) running and searching for foods
Grey Squirrel (Sciurus Carolinesis) running and searching for foods
Eastern Chipmunk (Genus Tamias, Tamias Striatus) probably running and playing with others
Song Sparrow (Family – Emberizdae, Genus – Melospiza melodia) appeared on the ground near short grass .probably searching insects
Conclusions:
The vertebrate and the invertebrate species made a symbiotic relationship. In my transect, song sparrow eats insects and use dead branch of plants to build the nest. Chipmunk can hide inside the bush to protect from bald eagle. Bald Eagle in the top of the food chain and carnivores. The vertebrate who live on vegetation or arthropod for energy are in level two. The plants on the surface and invertebrate microarthropods living in the soil are in level three. Bacteria identify in the soil and in the leaves are in level four. Bacteria and microarhropods helps plants to get nitrogen and other nutrition’s to grow and level two vertebrates depends on plants for their energy. If any one of the species eliminated from the niche will be harmful for the rest of the species.
MP
February 18th, 2015 – Invertebrates
Purpose: The purpose of this lab is to explore species (invertebrates) that live in transect five. We want to understand their classifications and importance of their role in this ecosystem.
Materials and methods: Last week, we placed our collected leaves in a Burlese Funnel underneath light. Invertebrate species moved to ethanol solution where they were preserved. We observed sample species from different phyla of invertebrates. First we observed acoelomates, pseudocoelomates and coelomaes and examined their features. Then, we observed five major classes of arthropods (Insect, diplopoda, crustacea, arachnida, chilopoda) which were preserved in different container. Lastly, we collect organism from preserved solutions underneath the Burlese Funnel and transferred into petri dishes for observation under a dissecting microscope. With the help of the invertebrate keys we identify five invertebrate species.
Data and Observations: we observed acoelomates, pseudocoelomates and coelomaes including the cross sections of an acoelomate and coelomate underneath the microscope. We observed Nemertea worms from the nematode phylum. their movement is slow back and forth movement. During movement their body part constricted in the middle and elongated at the front end to move forward.
Figure 1: Diagram of the cross section of coelomates
Figure 2: Observed cross section of coelomates under the microscope
Figure 3: insect
Figure 4: Diplopoda
Figure 5:Crustacea
Figure 6:
Figure 7:
Table 1 Showing common soil invertebrates:
Organism Length (mm) Number in Sample Description of Organism
1) Arthoropod, Insect 3 1 3 legs on each side of thorax; has front antennas; brown in color with black spots on thorax; three sections of body(largest)
2) Nematoda, Chromdaorea? 0.5 1 very tiny and thin; red in color; resembles a small string,(smallest)
3) Arthoropod, Insect 0.5 - 2 3 3 legs on each side of thorax; has front antennas; light purple in color with purple spots on thorax; three sections of body(most common)
4) Arthoropod, Insect 0.5 1 winged; larg wings the size of entire body; brown with antennas; three sections of body
5) Arthoropod, Insect 1 1 brown; wingless; structured similarly to Organism 3; dark brown; 3 sections to body
Conclusions: in our transect five Arthropod, insect are numerous in number and their size also largest among invertebrates. We found Nematoda is the smallest species in our specimens.
MP
Plantae and Fungi, 12th February 2015
Purpose: Examining and classifying the Plantae and fungi based on their vascularization, special structures and mechanism of reproductions. The study area (transect 5) is a small area but it has a variety of plants and fungi.
Materials and Methods: Five samples of plants were collected in a bag and some dead leaves also collected in another bag from the transect 5. After basic observation (like; size, shape, brunches etc.) the samples of plants were spliced and examined in the microscope to understand the vascular system. A ruler also used to measure the original size of the samples.
Sample from second bag (dead leaves) was placed on a Berlese funnel with a screen to hold the leaf in the funnel and to collect invertebrates. The Funnel was then taped to a 50ml tube filled with 25ml of 50/50 ethanol /water solutions. Then, the funnel was placed under the light with the foil cover on the top.
We also looked different samples of fungi including one sample from our agar plate under the microscope. We also studied PCR from last lab on a UV light box.
Picture1: weed
Picture 2 : Round weed
Picture 3:round bulb
Picture 4: bulb
Picture 5 : Grass
Conclusions: There are a lots of variety of plants in our transect. We collected only five. In summer there will be more plants in that area. Our next project will be study of invertebrate from samples of the funnel.
MP
February 4th 2015, Microbiology and identifying bacteria with DNA sequences
Purpose: In the lab, the bacteria from hey infusion culture which becames more moldy over weeks was studied in order to study bacteria. We used four plates containing agar and four plates with tetracycline antibiotics to grow bacteria. We want to understand the bacteria species, their antibiotic resistance and learn how to use DNA sequence to identify species. Archaea lives in extreme environment and are less likely to grow in agar plates.
Materials and Methods: We started with our eight plates with or without antibiotics. We observed the colonies and count the number of colony in each plates. Then we made we mount by placing a small sample from a colony on a slide along with a drip of water and covered with a cover slip. We also made gram stain by placing a small sample from a colony with a loop sterilized by a flame and labeled them. Then, we dried the slide on air by passing the slide on the top of a flame with bacterial smear side up. In the staining tray, we covered the bacterial smear with crystal violet for one minutes and then washed with water. Then, we covered the bacterial smear with Grams iodine mordant for one minutes and washed with water. Decolorized by flooding the bacterial smear with 95% alcohol for 20 seconds and rinsed gently. Then, covered the smear with safranin stain for 30 seconds and rinsed gently with water.
For the next lab, a colony of bacteria was placed in 100 microliters of water in a sterile tube and incubated for 10 minutes in a heat bath. After that we centrifuged for 5 minutes and added into the PCR tube with primer and placed them in PCR machine
Data and observations: The water level in hey infusion was little low and smells more moldy and a thick layer on the top.
Table 1: 100 –fold serial dilutions results
dilution Agar type Colonies counted Conversion factor Colonies/ml
10-3 Nutrient 1 lawn *103 n/a 10-5 nutrient 1500 *105 150000000 10-7 nutrient 24 *107 240000000 10-9 nutrient 0 *109 0 10-3 Nutrient+tet 480 *103 480000 10-5 Nutrient+tet 0 *105 0 10-7 Nutrient+tet 0 *107 0 10-9 Nutrient+tet 0 *109 0
Table 2: Bacterial characterization
Colony label Plate type Colony description Cell description Gram+ or Gram - Additional note Nutrient
107 Yellow
Light edge
circular Non motile circular -
spiral
Nutrient
107 Yellow
Circular
Fade at edge Non motile
Circular
Very tiny bundle together +
Nutrient+tet
103 Flat circular
Yellow
White circular Non motile
Inter twisted spiral -
Nutrient+tet
103 Yellow
Orange circular Multiple small circular dots +
Picture 1:growth in nutrient agar

picture2: growth in nutrient + tetracycline agar

Picture 3 : Gram stain slide showing gram positive coccus

We are Using PCR to determine tetracycline resistant gene on bacterial DNA. References: Connel, S. R. et al. (2003). Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrobial Agents and Chemotherapy47,12. Chopra, I & Roberts, M. (2002) Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance.US National Library of Medicine.'65(2).
MP
January 28th, 2015, Identifying Algae and Protists In hay Infusion with the help of dichotomous key
'Purpose: To Identify and understand eukarya especially unicellular Algae and protist under the microscope and learn how to use dichotomous key. We also prepared serial dilution of the sample of hey infusion to culture the organisms.
Materials and methods: We used hey infusion culture that we made in lab from the samples of transect 5.The hey infusion had rotten smell and very thin mold layer on the top. We took sample from the bottom of the jar and from the roots of the floating plants and made two slides to observe under the microscope. We examined microorganism in two different niches even though they are in the same jar. We observe and measure their size, shape and motility. For serial dilutions:
We labeled four tubes of 10 ml sterile broth with 10-2, 10-4, 10-6, and 10-8.
Obtained four nutrient agar and four nutrient agar with tetracycline plates and labeled them all with our names. We shacked the hay infusion culture to mix up all the organism and collected 100 microliter liquid from that solution to the 10 mls of broth in the tube labeled 10-.This is a 1: 100 dilution. Mixed it well. Than 100 microliter from tube 10-2 was added to tube labeled 10-4 and mixed it well. Repeated two more times to make the 10-6 and 10-8 dilutions. Then, added 100 microliter from each broth (10-2, 10-4, 10-6, and 10-8) to nutrient agar plate and nutrient agar plate with tetracycline. Carefully spreaded the sample on the plates and placed them to incubate.
Data and observations: We founded different sized and shaped organisms. Most of them are motile. From the bottom of the jar: Colpidium: 37.5 micrometer in length, motile, protist Chlamydomonas: 12 micrometer in length, motile, algae Peranema: 30 micrometer in length, motile, protist
From the rots of the floating plants:
Colpidium: 40 micrometer in length, motile, protist
Euglena: 12.5 micrometer in length, motile, protist
Peranema: 30 microliter in length, motile, protist

Conclusions and future directions: It is interesting to see the different organism under the microscope. Organisms vary based on the niches. Because different niche has different environment and variation in food sources. Next step will be to see the culture of those organism in agar plates.
MP
January 26th 2015. Observing a transect on American university to understand ecosystem
Purpose: To observe and understand the ecosystem between abiotic and biotic component of a small area and find out the organisms or species in that area. There should be lots of microorganisms and we should be able to culture them.
Materials and Methods: A 20 by 20 meter transect was observed and samples of biotic and abiotic component was collected. Approximate half of a sterile 50ml conical tube was filled with soil and rest of the container filed with berry, grass, weeds. In the lab we mixed our sample with 500ml deer park water and .1 gm of dried milk to prepare a hay infusion culture.
Data and Observations: Our transect was a portion of garden. Because of low temperature (35F), there were lots of dead leaf and plants.
Name of biotic and abiotic components
Biotic: Grass, Rose plants, Weed, Berry, Bush
Abiotic: Dead leaves, Soil, Stone, Snow, Wood chips
Conclusions: We will periodically monitor the portion of that garden and observe the change with the change of temperatures. In lab, we will culture the organisms to find out their characteristics.
MP