User:Madison Hayes/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| Line 27: | Line 27: | ||
The experiment concluded with identifying a couple of the bacteria strains of the agar plates. There was a noticeable difference between the plates that had been treated and the plates that had not in reference to how many colonies had grown on them. For future experiments the PCR reaction created during this one will be helpful in maybe determining the DNA sequence and from there analyzing how exactly certain bacteria had evolved to be resistant. This research could be helpful in trying to prevent deadly strains of antibiotic resistant diseases. | The experiment concluded with identifying a couple of the bacteria strains of the agar plates. There was a noticeable difference between the plates that had been treated and the plates that had not in reference to how many colonies had grown on them. For future experiments the PCR reaction created during this one will be helpful in maybe determining the DNA sequence and from there analyzing how exactly certain bacteria had evolved to be resistant. This research could be helpful in trying to prevent deadly strains of antibiotic resistant diseases. | ||
---- | |||
'''February, 10, 2014''' | '''February, 10, 2014''' | ||
Revision as of 14:44, 16 February 2014
February 16, 2014
Introduction:
The title of this experiment was Microbiology and Identifying Bacteria with DNA. The purpose of this experiment was to analyze the bacteria multiplied on the agar plates from the previous lab, determine their antibiotic resistance, observe the morphology, and set up a PCR reaction for the next experiment.
Materials and Methods: The materials for the experiment were as follows: the hay-infusion culture, the 7 agar plates from the previous week, cover slips, slides, microscope, oil immersion fluid, sterile loops, gram staining materials, bin, small tubes for PCR reactions, and micropipeters.
Procedures: 1) Notice if the hay-infusion has changed from the week before. 2) Count the number of colonies per agar plate and fill out the data chart. 3)Analyze the difference between the plates with and without antibiotics. How have the bacteria been affected? Record Data. 4)Make sure to label three colonies (two from non tetracycline plates) with a wax pencil. 5) Practice analyzing wet mounts with sample bacteria. 6) Create wet mounts of chosen, labeled colonies. 7) Identify bacteria, describe motility, draw a figure, and record. 8)Test if the samples are gram-positive or gram-negative. 9)Prepare PCR reactions of two of the samples for next weeks lab.
Observations and Data:
The hay-infusion jar looked slightly different from the week before. Quite a bit of the water had evaporated and the water had a clearer tint while some of the dirt appeared to have denigrated.
The following is the data chart for observing the colonies on the plates.

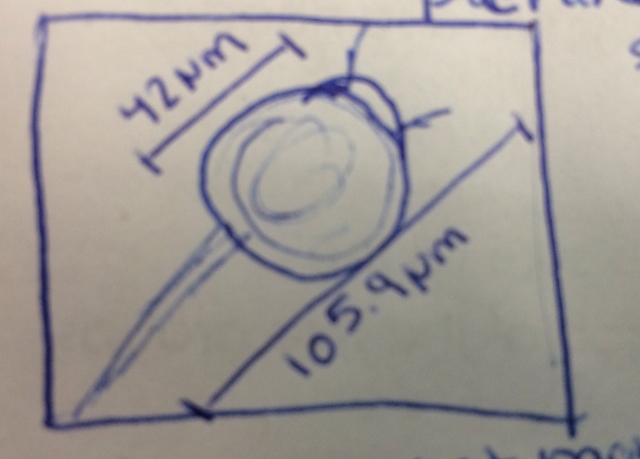
The next observations occurred when comparing the differences between the antibiotic resistance and the plates without antibiotics. The major differences between the plates were the number of colonies. There were significantly less colonies on the plates with antibiotics than the others. This indicates that there was less antibiotic resistant bacteria in the sample than resistant. The colors of and the bacteria also varied more in the plates without bacteria. The colors on the plates without antibiotics were orange, white, and blue while the tetracycline plates only had the orange colored colonies. Only one species was unaffected by the tetracycline,the convex, circular orange colonies. The bacteria chosen from the three colonies can be described as the following: (The diagrams were drawn based off of a 40x perspective.)
The bacteria resembles fusiform bacilli. It was not moving and did not have a particular arrangement. This sample was taken from an orange colony on the 10^-7 tetracycline plate. This bacteria tested gram-negative. 
This bacteria resembles spirilium. It was not moving and did not have a specific arrangement. This sample was taken from a white colony on the 10^-5 agar plate. This bacteria tested gram-positive. 
This bacteria resembles spirilium. It was not moving and did not have a specific arrangement. In contrast to the previous bacteria, it seemed longer and appeared more red in color. This sample was taken from an orange colony on 10^-7 agar plate. The bacteria tested gram-negative. 
Conclusion: The experiment concluded with identifying a couple of the bacteria strains of the agar plates. There was a noticeable difference between the plates that had been treated and the plates that had not in reference to how many colonies had grown on them. For future experiments the PCR reaction created during this one will be helpful in maybe determining the DNA sequence and from there analyzing how exactly certain bacteria had evolved to be resistant. This research could be helpful in trying to prevent deadly strains of antibiotic resistant diseases.
February, 10, 2014
Introduction:
The title of this experiment is Identifying Algae and Protists. The purpose of this experiment was to practice identifying groups of organisms using a dichotomous key and then put the practice to use identifying organisms in the hay infusion culture. The purpose is to identify the 6 organisms in the hay infusion sample created from soil and plant matter from transect five. The end of the experiment involved diluting the hay infusion culture and applying the dilutions to agar plates to set up for an experiment next week.
Materials and Method:
The following materials were utilized in the experiment: a two page dichotomous key, a sample of known organisms, microscope, slides, covers, transfer pipettes,protozoa, hay infusion culture (created last week). In order to prepare the dilutions for next week the following materials were needed: hay infusion culture, four tubes, 10 mL of water, micropipeters and tips, spreader, 4 agar plates without tetracycline, and three with tetracycline.
The procedures for the practice are as follows: 1. Carry the culture to the work station being careful not to jostle the environment in the jar. 2. Record your observations of the jar. 3. Take a sample from the jar and place a small drop on a slide and place a slide cover over it. 4. Characterize at least three different organisms from the slide. Draw pictures of each and measure the size. See if they can be identified with the key. 5. Repeat the steps with a sample from another area in the jar. 6. Obtain 4 tubes with 10 mL of water. Label the tubes with 2,4,6, and 8. 7. Find 4 nutrient agar plates and 3 nutrient agar plates plus tetracycline. Label the plates and add initials. 8. Shake the hay infusion mixture with the lid on and take 100 micro liters and add it to the tube labeled 10^-2. Mix tube. 9. Take 100 micro liters from the tube and add it to the next tube labeled 10^-4. Mix the tube. 10. Repeat two more times to create 10^-6 and 10^-8 dilutions. 11. For the nutrient agar plates, take 100 micro liters in 10^-2 and place on the surface of the nutrient agar labeled 10^-3. Repeat with the tetracycline plate labeled 10^-3. Carefully use a spreader to spread the sample around the plate. Make sure to use two seperate spreaders: one for the normal nutrient agar plates and one for the tetracycline plates. 12. Repeat the exact procedure for the remaining three tubes. The weakest dilution will only be placed on a nutrient agar plate not a tetracycline plate. 13. Allow the plates to sit at room temperature for a week.
Observations and Data:
The following describes the hay infusion culture unperturbed by motion. The dirt previously in the jar collected at the bottom. The water could be described as a murky clear mixed with some opaque white clouds. Some chunks of dirt floated on the top of the water instead of settling at the bottom. Pieces of the grass that had previously been in the infusion could not be seen; either the grass disintegrated or was buried among the dirt at the bottom of the jar. The jar had no particular smell that was noticeable.
The first niche from which the sample was taken was the top of the water. The second niche from which the sample was taken was from the bottom near the dirt. The samples were not taken from near any particular plant matter, but if it was the organisms could differ because organisms prefer different environments. Plant matter would offer different nutrients and a different environment to organisms leading to a different sample. Specific guesses of which on the dichotomous key it was are listed above the picture along with any specific notations needed not provided by the picture.
Niche 1--
Organism 1-Peranema sp Immobile
Organism 2- Colpidium sp Very mobile
Organism 3- Pandorna Circular whiplike motion
Niche 2--
Organism 1- Pandorina Immobile
Organism 2- Paramecium Bursaria Immobile
Organism 3- Chilomanas Immobile, especially dark spot in the center
Observations: Had the hay infusion been allowed to sit for another two months, I would hypothesize that the evidence of organisms would be more prevalent to the naked eye as the populations of organisms would be much greater. Another outcome could be that the organisms run out of nutrients and the organisms die. The slides would display unmoving organisms. The selective pressures that could have affected the compositions of the samples could have been the absorption of smaller organisms by their larger counterparts. A lack of nutrients could have killed off of some of the organisms that could not survive a nutrient drought.
The following picture describes the serial dilution process:

Conclusion:
The 6 organism samples from the hay infusion were discovered to be paranema, colpidium, pandorina, paramersium bursaria, and chilomanas. These organisms were found in the protazoa and paramecium categories. There was not a direct, straightforward hypothesis in this experiment. The dilutions to see if the bacteria grown in the infusion would be resistant to tetracycline were completed for the following week. Next week, the conclusions will most likely be that some of the bacteria will be resistant to the antibiotic because of the use of pesticides in the rosebed in which the bacteria was found.
January 31, 2014
Lab 1
Introduction: This lab was titled "Biological Life at AU". This lab was made up of two parts. The purpose of part 1 was to understand the cellular evolution of the volvacine line. The purpose of part two was to analyze the transects that will be assigned to us for the whole semester.
Procedure:
For part one the procedures are as follows: 1)Prepare slides of Chlamydomonas(make sure it is living) and analyze under microscope. 2)Add protoslo if necessary to properly view the alga. 3) Locate the conspicuous chloroplast and the the pyrenoid. 4)Repeat the steps examining volvox and gonium under a microscope. 5)For each sample record the data in the chart
For Part two the procedures were as follows: 1)Travel to assigned transect. 2)Describe the general characteristics of the transect. 3)List the abiotic and biotic factors of the transect 4)Using a 50mL conical tube take a soil and ground vegetation sample. 5)Weigh 10 to 12 grams of the soil and place in a plastic jar with 500 mL of deerpark water. 6)Add .1 grams dried milk and mix gently for 10 seconds. 7)Place in the back of the room with the top off.
Observations and Data
Part 1
Though evolution does not always lead to more complex organisms, that is the case with the volvacine line. The least complex of the three alga was the Chlamydomonas. The line evolutionarily advances as to gonium and then volvox. The colonies increase as from chlamydomonas to gonium and then to volvox.
Part 2
For this experiment transect 5 was analyzed. The transect is located in front of Hurst. The plot encompasses the following biotic factors: grass, rose bushes, bugs, and weeds. Soil, stone, bench, rocks, and a stone sign were observed abiotic factors. The location is in the middle of the quad and is relatively flat. The area is of high traffic so I would not predict too many larger animals will be seen in the transect. The vegitation in general was dead or winterized. More observations will be made throughout the semester.
Good start. Take care not to just re-write the directions from the protocol but to include real information. There could have been more information given and both parts explained in more detail. Include hypotheses and discussion of the text in red from the protocol. SK