User:Lisa White/Notebook/Biology 210 at AU: Difference between revisions
Lisa White (talk | contribs) |
Lisa White (talk | contribs) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 272: | Line 272: | ||
Above is a food chain of the animals that reside in T5. | Above is a food chain of the animals that reside in T5. | ||
== 0717, Development Lab == | |||
'''Purpose:''' To compare the stages and type of development for a chick, frog, human, and starfish. | |||
'''Materials and Methods:''' | |||
Incubated fertilized chicken eggs, slides of embryos in different stages, and research to figure out similarities and differences in the stages of development | |||
'''Results:''' | |||
[[Image:developmentt.png]] | |||
Above is a graph summarizing the results of the lab. | |||
== 0722, T5, DNA Bacteria Sequencing== | |||
'''Purpose:''' To identify the species of bacteria obtained in T5. | |||
'''Materials and Methods:'''The bacteria PCR reaction for 16S was sent out to an external laboratory to determine the DNA sequence of the bacteria culture. | |||
'''Results:''' | |||
Unfortunately, the results obtained from the lab did not produce any valid results. Instead a similar transect (transect 3) located very nearby, and of a similar ecosystem (tall grass) was used. The DNA bacterial sequence for this transect was the following: AAGATTAATACCCCATAATATTTTAAGTGGCATCACTTGAAATNGAAAACTCCGGTGGATAAAGATGGGCACGCTCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCTACGATCTTTAGGGGGCCTGANAGGGTGATCCCCCACACTGGTACTGAGACACGGACCANACTCCTACGGTAGGATCAGTGAGGAATATT | |||
By using the database NCBI Blast: Nucleotide Sequence, I was able to enter the sequence and find the bacterial organism that most closely matched the sequence. The result was Chryseobacterium, this bacteria is a soil dwelling bacteria, and has a gram stain negative. It can be inferred that this type of bacteria may also inhabit T5. | |||
Latest revision as of 10:42, 22 July 2014
0701, T5 collection and survey
Purpose: To make observe the characteristics of a particular transect deemed a "Wildlife Sanctuary" by the university and to and create a sample for future protist/bacteria studies.
Material and methods: The group observed both biotic and abiotic components of the 20x20 meter transect. Using a 50ml sterile conical tube, the group collect a sample of the area's foliage (including a bit of plant life and also local soil) that was representative of the ground surface of the transect. Back in the lab, 10-12 grams of the mixture was weighed and then placed in a plastic jar with 500 mls of deer park water. Then, .1g dried milk was mixed with the collected soil/foliage and gently mixed for 10 seconds. The top of the jar was the removed and labeled for future reference.
Data and observations: This particular transect was deemed a "Wildlife Sanctuary" of the university. It was located in front of Hughes Hall. Biotic components include Topography: small trees, small day lilies/ mondo grass "monkey grass", patches of moss ; Living organisms: mosquitoes, ants. Abiotic components': soil, few rocks, hot and humid environment, bench
Above is a picture of transect 5
0703, T5, identify protists and preparation for next week's lab
Purpose: To practice using a dichotomous key to identify unknown protists in the transect sample under the microscope, and to also examine algae from the transect under the microscope.
Materials and Methods:First students made a wet mount of the mixture from the transect and evaporated milk. Its size was recorded using the micrometer and corresponding conversion for the magnification. For each organism found, the student practiced using the dichotomous key to identify protists in the mixture. Students also noted the smell of the mixture and took samples from top, middle, and bottom (different niches) of the sample. Students drew drawings of organisms.
Data and observations: The mixture had a smell similar to that of feces. Within the mixture the heavier filaments like heavy bits of moss, foliage, and dirt sunk to the bottom while the top had minimal foliage filaments and a clear milky film. I was able to find Colpidium, chilomonas, and perarena. The Colpidium was fast and roundish shape and about 60 micrometers. The chilomonas was characterized by its double tail and was about 20 micrometers. The perarema was about 40 micrometers and colorless and vibrated.
Lab manual questions: Organisms may differ in kind being close to and from the plant matter. It is likely that those that associate with plants may consume plants or photosynthesize, while those farther away do not. Organisms found at the bottom in the part with the soil may be the ones accustomed to living in soil as their natural niche. If the Hay Infusion had been continued for another couple of months it is likely that more organisms would have been found. Hay Infusions increase the density of protists as the dried milk adds sugars for the protists to thrive and allowing the solution to sit for longer would allow for the density of protists to increase. A selective pressure in this particular hay infusion is a lack of food for protists that determines the carrying capacity for this particular niche. Furthermore, the amount of foliage or dirt present determines what specific protists were chosen and how well they can thrive in the environment chosen for them. The Chilomonas meets the requirements necessary for life. This protist conducts photosynthesis (uses and acquires energy), it is made up of a single cells, has distributed nuclei (processes information), and like all protists represent the ancient organisms from which other species have derived and they themselves represent products of evolution.
Pictured above is the mixture from which the wet mount was made.
Pictured above are drawings of the protists observed.
Materials and Method II Prokaryotic organisms and fungi will be observed next week. 4 tubes of 10ml sterile broth were labeled 10^(-2), 10^(-4), 10^(-6), 10^(-8), respectively and were filled with the 100mls of transect mixture. 4 nutrient agar and 4 agar plus tetracycline plates were then given given the corresponding 100mls of mixture. They will be incubated at room temperature for a week.
In this manner a serial dilution was conducted of the ecosystem mixture.
Labeling the petri dishes with the correct labels.
0708, T5, Identify bacteria with DNA sequence
Purpose: To observe and quantify and conduct gram stain tests on bacteria that grew in different serial dilution petri dishes, including those that grew in the presence of the antibiotic tetracycline. Also to prepare a PCR reaction in order to identify the types of bacteria by the 16S gene.
Lab book question: It is highly unlikely that Archaea species will grow on the agar plates because archaea typically live in extreme environments with very hot temperatures like volcanoes. The appearance or smell of a bacteria culture might change from week to week. This is because in a petri dish that has not yet harvested colonies of bacteria, it is likely that the smell will be lacking. When the bacteria begins to colonize and increase in growth it is likely that the smell will increase. Furthermore, if the type of bacteria changes from week to week in a sample then its particular scent will also change depending on what type of bacteria it is.
Material and Methods I: Gram Stain Procedure:
1. Sterilize loop over flame and scrape small amount from the surface. Mix it on a drop of water on slide. Circle area. Label slides.
2. Heat side over flame three times bacteria smear side up.
3. With a staining tray, cover smear with crystal violet for 1 minute
4. Rinse stain off using wash bottle with water
5. Cover smear with Gram's iodine mordant for 1 minute. Rinse gently
6. Flood smear with 95% alcohol for 10-20 seconds. Rinse. Decolorization occurs.
7. Cover smear with safranin stain for 20-30 seconds. Rinse.
8. Blot water with paper towel and air dry.
9. Focus on gram stained sample at a low magnification, observe under 40X and oil objective.
Materials and Methods II: Colony and Cellular Bacteria Observation : Choose 4 plates with distinctive characteristics. Note the smell and appearance of each and count the number of colonies on each plate. Use the dissection microscope to look at the shape of the colonies. Look at the stain underneath the compound microscope.
Do you see any differences in the colony types between the plates with vs without antibiotic?: The antibiotic has decreased the colony size of the bacteria by half. For example in 10^(-9) tet+ 35 colonies were counted and in dilution 10^-9tet- 61 colonies were counted. This indicates that for this particular species of bacteria 1/2 were antibiotic resistant and the other half were susceptible to the antibiotic. Overall the general trend was that tet killed about half of the bacteria cultures. 3 species of bacteria were able to grow in the presence of tet. The growth of bacteria on 10^-9tet was actually enhanced by tet as there are more orange colonies than in the 10^-9tet+ petri dish. It is not known what the effect of tet is on fungi growth since this experiment did not look into the growth of fungi. Tetracycline interferes with the bacteria's ability to produce proteins that are essential to them. Without these proteins the bacteria grow in large numbers (Princeton).
Data and observations:
Number of colonies & the number of colonies per ml.
Above is a photo of the cultures of bacteria.
This table describes the bacteria on a colony and cellular level.
Above are rudimentary drawings of the bacteria types examined under the microscope at 100x
Material and Methods III: PCR Set up for 16s Sequencing
1. Select 3 plates with the best characterization & label each sample
2. Transfer single colony to 100 micoliters of water in a tube
3. Incubate at 100 degrees C for 10 mins
4. Centrifuge boiled samples for 5 mins at 13,400 rps
5. During centrifugation, add 20 micro milers of primer/water mixture to a labeled PCR tube and mix
6. Transfer 5 micro liters of supernatant to the 16s PCR reaction and place in PCR machine
7. Next week the PCR products will be put in a agarose gel.
Works Cited:
Tetracycline. (n.d.). Princeton University. Retrieved July 8, 2014, from https://www.princeton.edu/~achaney/tmve/wiki100k/docs/Tetracycline.html
0710, T5, Characterize Plants and Prepare Berlese Funnel to Collect invertebrates
Purpose:To characterize plants found in transect area and to prepare a berlese funnel to collect invertebrates for next week's lab.
Materials and Methods:
1. Collect 5 live plant samples from transect. 2. Collect leaf litter for Berlese Funnel. 3. Describe the 5 plants and their location. 4. Use info to identify the major group from the table above and reference additional resources to determine the genera for each of the 5 samples.
Data:
Table 1
Above are the observations and genera conclusions made about the types of plants found in the transect.
Above is a photo of the leaves classified in Table 1.
Sporangia are small black circular structures in fungi that contain spores that allow the fungi to reproduce.
Purpose II: To observe and identify fungi under the dissecting microscope. Additionally fungi were observed under a dissecting microscope. There were 3 groups observed Zygomycota which has a thick wall. Ascomycota is in the form of a U shape. The Basidiomycota can be recognized because of their large mushroom like caps.
Results: The fungi observed under the microscope was Rhizopus or bread mold. It consists of sporangia which are small black dots and mycelium which are the lines connecting it together. It appears to be a fungus because it contains the reproductive parts characteristic of Rhizopus (the sporangia and mycelium).
Above is a rudimentary drawing of Rhizopus
0715, T5, To identify invertebrates found in the transect
Purpose: To observe invertebrates and to identify invertebrates found in the transect.
Methods and Materials I:
1. Observe planaria that has been fed egg yolk using a dissecting scope. Note its movement. *Note the planaria observed were not alive. Then look at the cross section slide of planaria under the microscope. The digestive track is stained.
2. Observe the live nematodes and note their movement next observe their corresponding cross section slide.
3. Next observe the coelomate, Annelidia.
Results
1. The planarium's movement was not observed, however, planarium moves by stretching itself in an almost slithering way. Planarium is a flatworm with bilateral symmetry and for this reason it makes sense that it would creep in this way.
Drawings of planarium.
2. The nematodes had a slithering creeping motion. Like a worm, the nematode has no appendages because it is cylindrical and smooth it is able to move in such a motion.
3. The movement of the Annelidia was not observed. However, Annelidia moves segment by segment by inching forward. It's movement is like this because it has no appendages, but is muscle-like
Drawings of Annelidia and nematode.
Methods and Materials II
1. Break Berlese setup and pour top organisms into one petri dish and the bottom organisms into another.
2. Examine both under a dissecting microscope.
3. Use a key to identify the phylum and class of the invertebrates.
Note: worms will dry up rapidly, and insect grubs will not be captured
4. Select 5 invertebrates and identify them and measure them.
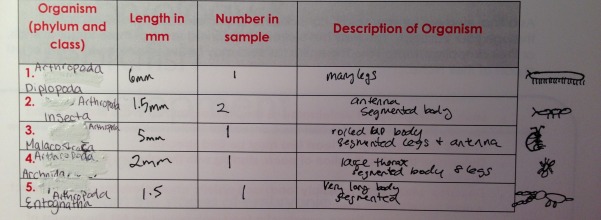
Results
Above is a chart summarizing the invertebrates fount in T5.
The termine #2 was the most common. The size range of all arthropods was 1.5-6mm. The largest was the millipede #1 the smallest was the springtail #6 and termite #2. All of these organisms were found at the pellet of the leaf litter.
Materials and Methods III
1. Consider 5 vertebrates that inhabit T5, determine the classification of each, what biotic and abiotic components of T5 benefit the species. 2. Construct a food web based on all the groups of organisms observed. Describe how these organisms represent the ecological concepts of community, carrying capacity, and trophic levels.
Results
1. Sparrow
Classification: Animalia, Chordata, Aves, Passeriformes, Passeroidea, Passeridae, Passer
Biotic: Annelidia for food and other invertebrates, tree for shelter
Abiotic: Sticks for creating nest
2. Robin
Classification: Animalia, Chordata, Aves, Passeriformes, Turdidae, Turdus, T. migratorius
Biotic: Annelidia for food and other invertebrates, tree for shelter
Abiotic: Sticks for creating nest
3. Squirrel
Classification: Animalia, Chordata, Mammalia, Rodentia, Sciuromorpha, Sciuridae, Sciurus
Biotic: Nuts from tree for food, insects, tree for shelter, grass for hiding
Abiotic: trashcan for food
4. Wood Frog
Biotic: Consumes arthropods, and slugs snails and worms
Abiotic: Rocks for hiding and shelter
Classification: Animalia, Chordata, Amphibia, Salientia, Anura, Ranidae, Lithobates, L. sylvaticus
5. Eastern Chipmunk
Biotic: Trees (Likes to live in wooded areas), shrubs for shelter. Eats bulbs, seeds, fruits, nuts, green plants, mushrooms, insects, worms, and bird eggs.
Abiotic: rocks for shelter
Classification: Animalia, Chordata, Mammalia, Rodentia, Sciuridae, Marmotini, Tamias, T. striatus
Above is a food chain of the animals that reside in T5.
0717, Development Lab
Purpose: To compare the stages and type of development for a chick, frog, human, and starfish.
Materials and Methods:
Incubated fertilized chicken eggs, slides of embryos in different stages, and research to figure out similarities and differences in the stages of development
Results:
Above is a graph summarizing the results of the lab.
0722, T5, DNA Bacteria Sequencing
Purpose: To identify the species of bacteria obtained in T5.
Materials and Methods:The bacteria PCR reaction for 16S was sent out to an external laboratory to determine the DNA sequence of the bacteria culture.
Results:
Unfortunately, the results obtained from the lab did not produce any valid results. Instead a similar transect (transect 3) located very nearby, and of a similar ecosystem (tall grass) was used. The DNA bacterial sequence for this transect was the following: AAGATTAATACCCCATAATATTTTAAGTGGCATCACTTGAAATNGAAAACTCCGGTGGATAAAGATGGGCACGCTCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCTACGATCTTTAGGGGGCCTGANAGGGTGATCCCCCACACTGGTACTGAGACACGGACCANACTCCTACGGTAGGATCAGTGAGGAATATT
By using the database NCBI Blast: Nucleotide Sequence, I was able to enter the sequence and find the bacterial organism that most closely matched the sequence. The result was Chryseobacterium, this bacteria is a soil dwelling bacteria, and has a gram stain negative. It can be inferred that this type of bacteria may also inhabit T5.