User:Gabriel H Gan/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| (80 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
TA Alyssa | TA Alyssa Pedersen | ||
MW 5:30-8:40PM | MW 5:30-8:40PM | ||
'''Lab 1: Observing a Niche at AU''' | '''Lab 6: Analysis of Bacteria Sequencing''' 7/28/14 | ||
'''Introduction''' | |||
In Lab #3, two colonies of bacteria were chosen to be set up for PCR in the hopes of having products that can be sent out for 16s sequencing. This post will present and discuss the results of Transect #3's actual bacterial sequencing results. | |||
Bacteria are a very diverse domain, and different bacteria have their own specific enzymes (Bentley et al., 2014). The enzymes are specific due to the fact that the DNA sequences are different for the genes that code for the enzymes. PCR with a special primer that is specific for these genes can be employed to identify species, as well as show relatedness between different species (Bentley et al., 2014). | |||
---- | |||
'''Materials and Methods''' | |||
Back in Lab #3, the selected bacteria colony was transferred to a 100 microliter tube and incubated at 100 degrees C for 10 min. Next, the tube was centrifuged for 5 minutes at 13,400 rpm. A separate, labeled tube was obtained and 20mL of primer/water mixture with a PCR bead was added. 5 micro liters of the supernatant of the boiled/centrifuged samples was placed in the 16s PCR reaction, and the tube was placed in the PCR machine. | |||
The PCR products were run on an agarose gel by the lab TAs. The PCR product was then sent out for sequencing. | |||
The 16s sequence was analyzed using NCBI nucleotide blast to determine its identity. | |||
All procedure explained above was followed per the 2014 Biology 210 lab manual by Bentley et al. | |||
---- | |||
'''Results''' | |||
The 16s sequence that came back from the lab was as follows: | |||
AAGATTAATACCCCATAATATTTTAAGTGGCATCACTTGAAATNGAAAACTCCGGTGGATAAAGATGGGCACGCTCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCTACGATCTTTAGGGGGCCTGANAGGGTGATCCCCCACACTGGTACTGAGACACGGACCANACTCCTACGGTAGGATCAGTGAGGAATATT | |||
NCBI blast identified this sequence at 95% for Chryseobacterium. The corresponding accession number is JQ660730.1 | |||
From the results of the previous Bio 210 classes this year, 10 more sequences were identified using NCBI blast. These included Chryseobacterium, Flavobacterium, Pseudomonas, Flectobacillus, Sphingobacterium, and Aeromonas. | |||
Looking back at our identifications from lab 3, the samples we identified were gram + and rods. This is different than the BLAST identified bacteria, as the ones investigated (chryseobacteria, flavobacteria, pseudomonas) are all gram - and rod shaped. | |||
---- | |||
'''Tables and Graphs''' | |||
Table 1: Bacteria Identified in Transect #3 | |||
[[Image:Ganbacteriablast.png]] | |||
The table above shows a sample of bacterium identified from transect #3 using NCBI Blast. The sequences identified came from both our group's sample as well as samples from previous BIO 210 classes. Once identified, the above characteristics of the bacterium were determined (Im, 2011; Genoscope, 2012; Todar, 2012). In the 11 sequences analyzed, 5 were ''Chryseobacteria''. | |||
---- | |||
'''Discussion:''' | |||
The purpose of this lab was to identify the bacteria present in transect #3. Initally, our group discussed that we would expect a large variety of bacteria, as bacteria is so abundant and diverse. I was somewhat surprised to see that Chryseobacterium was so common and represented a large number of the sequence results. Chryseobacterium is abundant in soil, which our transect is largely composed of. ''Chryseobacterium'', ''Flavobacterium'', and ''Pseudomonas'' are all also abundant in water, so it is possible it has ended up in our non-aquatic transect in run-off (Im, 2011; Genoscope, 2012; Todar, 2012). | |||
One important limitation is that the samples provided to us from previous classes were from different seasons. Our samples were from the summer. Different organisms may be present in different seasonal conditions. | |||
The difference in our visually identified bacteria from lab #3 and the BLAST results may be due to lack of experience. Our group judged bacteria with any type of stain as gram +; we now know that the difference between gram + and - is not the presence of stain but rather the color (purple vs. pink). | |||
---- | |||
'''References''' | |||
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C. | |||
"Flavobacterium" Genoscope. 2012. Centre National de Sequencage. (25 Jul 2014). <http://www.genoscope.cns.fr/spip/-Flavobacterium,511-.html> | |||
Im, WT et. al. 2011. Chryseobacterium ginsenosidimutans sp. nov., a bacterium with ginsenoside-converting activity isolated from soil of a Rhus vernicifera-cultivated field. Int J Syst Evol Microbiol. 61: 1430-5 | |||
Todar, Kenneth. 2012. Todar's Online Textbook of Bacteriology. University of Wisconsin: Madison, WI. | |||
---- | |||
---- | |||
'''Lab 5: Analysis of Invertebrates and Vertebrates in Transect #3''' 7/14/14 | |||
'''Introduction''' | |||
In this lab, the invertebrates collected from transect #3 were analyzed. There are 14 different phyla of invertebrates, all ranging in size, shape, and complexity (Bentley 2014). Porifera, or sponges, are among the simplest invertebrates, while Platyhelminthes are more complex (Bentley 2014). Most of the invertebrate findings from transect #3 are arthropods, which consist of adult insects and their larvae (Bentley 2014). Invertebrates vary in their symmetry, embryonic tissue layers, and digestive development (Bentley 2014). The invertebrates collected from transect #3 are diverse and exemplify the variety of the invertebrate group. | |||
---- | |||
'''Materials and Methods''' | |||
All procedures were performed using the Bio 210 Lab Manual. | |||
First, the prepared slides and samples were observed under the microscope and through the naked eye to gain experience in identifying, analyzing, and classifying invertebrates. The nematoda sample was observed for structure and movement. Some of the worms and samples arrived dead, so they were not observed. | |||
Next, the Berlese setup was broken down to obtain the invertebrates that should have collected in the ethanol. The collection tube was poured into petri dishes for analysis, but no invertebrates were found. As none were found, the funnel leaf litter contents were dumped into a large tray. Three invertebrates were found alive and placed in the ethanol collecting tube to be analyzed with microscopy. Two samples were borrowed from other transect groups (transects #1 and #5) as our group only found 3 organisms. | |||
Each sample was placed in a depression slide. Both a dissecting microscope and compound microscope were used to observe sample. The samples were measured and a description of the organism was recorded in the lab notebook. The dichotomous key distributed in lab as well as Google image searches were used to identify the organisms. | |||
The last part of the lab was research of vertebrates using the textbook and internet sources. From this, a food web was created. | |||
---- | |||
'''Results''' | |||
The only worm movement examined was of the nematoda because the other samples were dead. Many nematodes were visible on the slide and there was variation in their size to some degree. The nematoda wiggled longitudinally. This fits their long slender bodies that have limited muscles to produce an undulating movement. | |||
Figure 1: Nematoda Sample Slide | |||
[[Image:Gannematoda.png]] | |||
Figure 1 shows nematoda as visualized through a compound microscope. | |||
In transect #3, our group identified a biting louse, a spider of unknown type and a armadillidiidae (pill bug). Below are pictures and descriptions of each organism. The data is organized into table 1 in the "Tables and Graphs" section. | |||
Figure 2: Organism #1: Biting Lice | |||
[[Image:Ganlice.png]] | |||
One was found in transect #3. It had six legs, antennae, hairy body, hairy legs, and was 1.125mm. | |||
Figure 3: Organism #2: Spider of Unknown Type | |||
[[Image:Ganspider.png]] | |||
One of these spiders was found in transect #3 leaf litter. The organism appeared hairy, with a large round thorax, brown in color, with 8 legs. It measured 0.625mm. | |||
Figure 4: Organism #3: armadillidiidae (pill bug) | |||
[[Image:Ganarmadillidiidae.png]] | |||
One was found in transect #3 leaf litter. It had about 10 legs, a segmented exoskeleton, and antennae. The sample was too large to measure via ocular micrometer. | |||
Figure 5: Organism #4: Biting Lice | |||
[[Image:Ganbitinglice.png]] | |||
This sample came from transect #5. It had six legs, hairy antennae, and a head with a long striped body. It measured 1.95mm. | |||
Figure 6: Organism #5: Millipede | |||
[[Image:ganmillipede.png]] | |||
This sample came from transect #1. It had a segmented hairy body with many legs. It measured 3.5mm. | |||
---- | |||
'''Tables and Graphs''' | |||
Table 1: Invertebrate Organisms found in Transect #3 Leaf Litter | |||
[[Image:ganinvertebratetable.png]] | |||
Above are the organisms found in the transect #3 leaf litter obtained on 7/7/14. Note that the last two rows are of organisms from different transects. | |||
Table 2: Expected Vertebrates of Transect #3 | |||
[[Image:Gan5Vertebrates.png]] | |||
The above organisms are a prediction. The only vertebrate that has been noticed in the transect are squirrels. | |||
---- | |||
'''Discussion:''' | |||
The purpose of this lab was to identify and study different phyla of invertebrates in transect #3. | |||
The size range is from 0.625 mm (the spider) to larger than the ocular micrometer (Armadillidiidae). It is impossible to determine exactly what kinds of organisms are most common in leaf litter because we only obtained one of each sample. From the phyla, it appears that arthropoda are common in leaf litter. Although we identified many arthropoda, there are other invertebrates that are | |||
vital to the transect. | |||
The louse from transect #3 appeared different than the louse we borrowed from transect #5. While they both had 6 legs and hairy bodies, the louse from transect #5 had a different coloration and stripe pattern. The dichotomous key identified both organisms as biting lice, but that is just a broad category. There are over 3000 known species of lice, so it is most likely the case that each organism is a different species (Christensen 2005). | |||
It is possible that no invertebrates were observed in the ethanol collecting tube because our leaf litter sample had too much soil in it. The thicker sample may have prevented the passage of organisms down the funnel, through the screen material, and into the collecting tube. | |||
The identification of the sample organisms may be incorrect due to limited experience identifying invertebrates as well as the fact that the dichotomous key had a limited number of species it led to. | |||
Five vertebrates that may inhabit the transect are the Eastern Gray Squirrel, the Eastern Garter Snake, Raccoon, the American Crow, and the American Robin. (Metropolitan Naturalist). The squirrel was observed in the transect. The four other species were predicted inhabitants after reviewing web sources on the animals native to the D.C. area. The scientific names and classification of each vertebrate show there are mammals, reptiles, and aves, but that they differ from order down, with exception to the two birds that both share the order Passeriformes. The phylum through species categorization of each vertebrate is located in Table 2. | |||
Biotic and abiotic characteristics of the transect can benefit each species. Squirrels live in leafy nests on tree branches, and transect #3 has two large trees for squirrels to make these nests (Metropolitan Naturalist). Garter snakes eat worms and even mice, which are most likely present in the transect (Metropolitan Naturalist). Raccoons should be visitors of the transect due to the proximity of garbage cans, as they eat trash, as well as due to the smaller animals in the transect which they prey on (Metropolitan Naturalist). The transect provides tall trees for crows to perch atop, and since crows eat trash, there is trash littered by humans in the transect as people walk on the path through it. The transect can support robins because it has plenty of branches to make nests and the grass/dead leaves create a good camouflage environment for them. | |||
The following food web has been constructed to show the relationships between transect #3 organisms. | |||
Figure 7: Transect #3 Food Web | |||
[[Image:GanFoodWeb.png]] | |||
Food web shows energy flow between bacteria, plant matter, invertebrates, and vertebrates. This diagram is just of the identified organisms of transect #3; our group acknowledges that there are many, many more organisms that we did not identify and are not on this web. | |||
The concept of community states describes when within a particular area, populations of different species interact (Freeman 2014). The food web shows the different organisms that live in the transect together that interact through consuming each other. Carrying capacity is the maximum population that an environment can sustain (Freeman 2014). The resources in transect #3 are limited in terms of food and housing; this affects the number of each species and the balance of organisms in the transect. Trophic levels refer to the feeding levels in an ecosystem (Freeman 2014). The food web shows the different tropic levels of transect #3, with the dead animals and leaves as well as live leaves and flowers in the first trophic level, all the way to the vertebrates, which are in the highest trophic level. | |||
- | |||
'''References''' | |||
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C. | |||
Christensen, Karin. "External Parasites". ''The Biology of the Goat.'' 2005. (16 July 2014). <http://www.goatbiology.com/lice.html> | |||
"Some Animals of the Metro Area". ''Metropolitan Naturalist''. (16 July 2014). <http://metro-naturalist.com/Animals/Fauna_Menu.html> | |||
---- | |||
---- | |||
'''Lab 4: Studying Plants and Fungi in Transect #3''' 7/9/14 | |||
'''Introduction''' | |||
Today's land plants are thought to have evolved from bryophytes, non-vascular mosses (Bentley 2014). These mosses and simple plants evolved when water space became competitive and land space was wide open. Plants evolved to tolerate the temperature differences on land, water and nutrient access issues, as well as male and female gametic interaction (Bentley 2014). Tracheophytes diverged from the bryophytes in the Devonian period and radiation occurred; in the more recent Cretaceous period, Angiosperms (flowering plants) radiated into many niches (Bentley 2014). | |||
In this lab session, the plants and fungi on transect #3 were analyzed. Special note was taken of the vascularization, specialized structures, and reproduction anatomy of these plants. | |||
---- | |||
'''Materials and Methods''' | |||
The first procedure performed was collection of plant samples from the transect. A leaf litter sample consisting soil and dead leaves was collected into a large Ziploc bag to be used in the Berlese funnel. Next, a sample was taken from five plants in the transect in the form of a flower or leaf. | |||
Back in the lab, we first practiced analyzing known plant and fungi samples with both the naked eye as well as through compound and dissecting microscopy. Seeds were dissected to identify the different parts. After practicing, we analyzed the five plant transect samples and recorded notes on vascularization, specialized structures, and reproduction. | |||
The last procedure was setting up the Berlese funnel for invertebrate collection. A funnel with a screen at the base was connected to a flask filled with preservative. A light source was placed above the funnel. The soil and leaf litter sample was placed in the funnel. (Bentley 2014). | |||
---- | |||
'''Results''' | |||
No seeds were obtained from the transect. | |||
The leaf litter sample was obtained from the main bed of the transect around the front tree and low bushes. See picture below of leaf litter collection site. | |||
Figure 1: Leaf Litter Sample Collection Site | |||
[[Image:Ganleaflittercollectionsite.png]] | |||
The leaf litter sample for the Berlese funnel was collected next to a bush and a tree and consisted of dry leaves and dirt. | |||
The plant samples were obtained from a wide variety of plant life in the transect. The updated transect map below shows the sample number and location it was collected from. A leaf or flower was picked from each plant as a representative sample. All of the plants had vascular structures. Our group did not obtain any seeds from the transect, so we cannot be sure about plant vascularization. | |||
Sample #1 and #5 are of angiosperms. Angiosperms are flowering plants that are a group of seed plants (Freeman 2014) . Sample #1 was a white, conical flower that was approx. 6cm in diameter. This flower came from a tall green bush. Sample #5 was a yellow, five-petal flower that was approx. 5 cm in diameter and came from a short green bush. | |||
Table #1 in the "Tables and Graphs" section shows the characteristics of all five transect plant samples. | |||
Figure 2: Map of Transect #3 with Plant Sample Locations | |||
[[Image:Ganupdatedsamplemap.JPG]] | |||
The figure above is an updated map of the transect with the numbers 1-5 to denote the locations the plant samples were taken. | |||
Figure 3: Plant Sample #1 | |||
[[Image:ganplant1.png]] | |||
This plant is located near light post C; it is a green bush with white flowers. | |||
Figure 4: Plant Sample #2 | |||
[[Image:ganplant2.png]] | |||
This plant is located in the front of the transect near the bench; it is a tree with white flowers. | |||
Figure 5: Plant Sample #3 | |||
[[Image:ganplant3.png]] | |||
This plant is located in the middle of the transect; it is a maple tree. | |||
Figure 6: Plant Sample #4 | |||
[[Image:ganplant4.png]] | |||
This plant is located in the middle of the transect; it is a tall green bush. | |||
Figure 7: Plant Sample #5 | |||
[[Image:ganplant5.png]] | |||
This plant is located near light post A; it is a short green bush with yellow flowers. | |||
Figure 8: Flowers and Leaf Samples from Plant Samples | |||
[[Image:Gansamples1to5.png]] | |||
The picture above depicts the five samples obtained. Starting from the left, they are ordered 1-5. | |||
Figure 9: Fungi observation | |||
[[Image:ganbreadmold.png]] | |||
The fungi observed was of ''Rhizopus stolonifer'', commonly known as black bread mold. | |||
Figure 10: Sketch of Fungi Under Microscope | |||
[[Image:Ganbreadmoldimage.png]] | |||
The above sketch is of ''Rhizopus stolonifer'', commonly known as black bread mold. | |||
---- | |||
'''Tables and Graphs''' | |||
Table 1: Characteristics of Transect #3 Plant Samples | |||
[[Image:Gansampletable.png]] | |||
Table 1 shows the characteristics of the five plant samples obtained from transect #3. Each plant was similar in that they were vascularized, but they each had different specialized structures. Classification on reproduction was estimated using the monocot vs. dicot table in the lab manual to characterized observed morphologies as no seeds were available for analysis. | |||
---- | |||
'''Discussion:''' | |||
Through this lab, we sought to identify and analyze plant samples from transect #3. Bushes and trees are vascular and seed producing. In the transect, we saw diversity in the leaf type and size, as well as in the flowers. The leaves varied in shape from oval to multi-pronged and they were of different shades. Some had more of a wax covering than others, and some had spiked ends, presumably for defense and protection. The flowers varied from conical shaped to petaled, and from white to yellow. | |||
Without seeds to examine, we had to make educated guesses on whether the plant samples were monocot or dicot. Our group acknowledged that analysis of cotyledons in seeds would have been preferable to determine whether the samples are monocot or dicot. From the lab manual table, monocots usually have long narrow leaves with parallel veins and dicots have broader leaves with a network of veins (Bentley 2014). It was also possible to base the decision on floral arrangement, as monocots usually have floral parts in multiples of three and dicots in multiples of either four or five (Bentley 2014). Sample #2 and #4 had parallel veins and a more narrow leaf structure that correlates with monocots. In contrast, sample #3 was of broad shape and had a branch like vein structure. Samples #1 and #5 both had petals in multiples of four or five, which correlates with dicot structure. | |||
Fungi sporangia are cells that holds spores until they are mature and ready to be dispersed (New Brunswick Museum). Sporangia are important because they cane be the site of meiosis or mitosis and that they are responsible for spreading fungi. Fungi can reproduce asexually by means of sporangia (New Brunswick Museum). The samples observed under the microscope appeared to be fungi. One was a Rhizopus stolonifer (bread mold), which is a zygomycota. It appeared to be a zygomycota due to the thick walled zygospore present (Bentley 2014). There are many types of fungi, but most are filamentous and fungi have cell walls and spores (Bentley 2014). The samples observed under the microscope had these features. | |||
---- | |||
'''References''' | |||
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C. | |||
"Fungi Reproducing Asexually By Means on Sporangia". Natural History of Fungi. N.d. New Brunswick Museum. (13 July 2014) <http://website.nbm-mnb.ca/mycologywebpages/NaturalHistoryOfFungi/AsexualSporangia.html> | |||
---- | |||
---- | |||
'''Lab 3: Analysis of Bacteria in Transect #3''' 7/7/14 | |||
'''Introduction''' | |||
This lab focuses on the study of the bacteria found in transect #3. There are different groups of bacteria, including Proteobacteria, Chlamydiae, Actinobacteria, Spirochetes Cyanobacteria, and Firmicutes (Bentley 2014). Biologists classify bacteria on morphology, and examine characteristics such as form, elevation, margin, size, and stain characteristics (Bentley 2014). A positive gram stain yields bacteria that remains violet after preparation due to the thick peptidoglycan wall, while negative gram stains do not hold the dye (Bentley 2014). | |||
In this lab, the agar plates prepared by serial dilution in lab 2 were analyzed by regular observation with microscopy as well as with gram staining. At the end of lab, PCR was set up for 16s sequencing to be run through gel next lab class. | |||
I do not believe any Archaea species will have grown on the agar plates. Since Archaea tend to grow in extreme environments, the petri dish with agar will be no match. The room temperature petri dish is much cooler than a hot spring, one environment in which Archaea thrive (Bentley 2014). | |||
I predict that the plates with nutrient agar only will have more diverse and higher numbers of bacteria colonies in comparison to the nutrient agar + tetracycline plates. If tetracycline is effective against the transect 3 bacteria, then there will be fewer colonies observed on the corresponding agar plates. | |||
---- | |||
'''Materials and Methods''' | |||
First, all six petri dishes were placed on the bench for visual inspection with the naked eye. From this, it was possible to gather general information about the number of colonies, pattern, colors, and shapes. | |||
Next, wet mounts were prepared from two of the nutrient agar petri dishes and one of the tetracycline petri dishes. These wet mounts were examined under the compound microscope at 1000x using oil. | |||
After, gram stain slides were prepared from the same colonies used for the three regular wet mounts. The colony samples were heat fixed to the slide, stained with crystal violet, and set with Gram's iodine mordant and safranin stain. Observations were recorded under 400x and 1000x. | |||
The final procedure was the PCR setup for 16s sequencing. Bacteria from each of the three colonies used in the wet mount and gram stain was incubated and then centrifuged. Samples from these three tubes were then mixed with a PCR bead and placed into a smaller tube with 16s PCR reaction liquid (Bentley 2014). | |||
'''Results''' | |||
Our group briefly glanced at the Hay Infusion and there was nothing to report on any changes in appearance. The Hay Infusion was not opened so no observations were noted on changes in smell. | |||
There were differences noted between the plates with antibiotic and the plates without antibiotic. All of the tetracycline plates had yellow colonies. In contrast, the nutrient agar plates varied in color, from red to white to yellow colonies. In general, there was more variation noted on the nutrient agar plates, including different edges on the colonies and the presence of a small lawn on the 10^-5 plate, as well as more colonies. The tetracycline plate colonies appear to have entirely even margins. | |||
Our transect group chose to take samples of colonies from the 10^-9 agar+tet, 10^-9 agar, and 10^-5 agar dishes. See figures 3, 4 and 5 for images of these selected colonies. From these samples, no bacteria was motile. The 10^-9 tetracycline + agar and 10^-5 agar samples appeared gram positive, while the 10^-9 agar was gram negative. In regards to bacterial shape, the 10^-5 agar sample appeared to be coccus shaped while the 10^-9 tetracycline + agar sample was bacillus shaped. See images and tables below. | |||
Figure 1: 100 fold Dilutions Petri Dishes (First Four) | |||
[[Image:Gan petri1.png]] | |||
These were grown with nutrient agar. Bacteria source is transect 3 hay infusion culture. | |||
Figure 2: 100 fold Dilutions Petri Dishes (Second Four) | |||
[[Image:Gan petri2.png]] | |||
These petri dishes contained nutrient agar and tetracycline. Bacteria source is transect 3 hay infusion culture. | |||
Figure 3: 10^-5 Nutrient Agar Petri Dish | |||
[[Image:Gan10-5.png]] | |||
This dish was chosen for microscopy, gram staining, and PCR. The sample was taken from one of the larger yellow colonies. | |||
Figure 4: 10^-9 Nutrient Agar Petri Dish | |||
[[Image:Gan10-9.png]] | |||
This dish was chosen for microscopy, gram staining, and PCR. The sample was taken from the large red colony. | |||
Figure 5: 10^-9 Nutrient Agar + Tetracycline Petri Dish | |||
[[Image:Gan10-9T.png]] | |||
This dish was chosen for microscopy, gram staining, and PCR. The sample was taken from a larger yellow colony. | |||
Figure 6: View of Gram Stained Sample from 10^-9 Tet Petri Dish | |||
[[Image:Gan gramstain1.JPG]] | |||
This image shows the gram positive colony of bacteria from the 10^-9 agar and tetracycline petri dish at 1000x. | |||
Figure 7: View of Gram Stained Sample from 10^-9 Tet Petri Dish | |||
[[Image:Gan gramstain2.JPG]] | |||
This image shows the gram negative colony of bacteria from the 10^-9 nutrient agar petri dish at 1000x. The red seen in the photo is from the wax pencil, there is no stained bacteria present. | |||
Figure 8: View of Gram Stained Sample from 10^-5 Petri Dish | |||
[[Image:Gan gramstain3.JPG]] | |||
This image shows the gram positive colony of bacteria from the 10^-5 nutrient agar petri dish at 1000x. | |||
---- | |||
'''Tables and Graphs''' | |||
Table 1: Results of Serial Dilutions in Nutrient and Nutrient + Antibiotic Agar Plates | |||
[[Image:100foldserialdilutiongan.png]] | |||
This table shows data from colonies counted on plates with different dilutions. Nutrient agar plates had more bacterial colonies observed. | |||
Table 2: Bacteria Characterization of 10^-9 Tet, 10^-9, and 10^-5 Plate Colonies | |||
[[Image:Bacterial Characterization Gan.png]] | |||
The data presented above is from the analysis of gram staining the three colonies selected from 10^-9 agar and tetracycline, 10^-9 nutrient agar, and 10^-5 nutrient agar. | |||
---- | |||
'''Discussion:''' | |||
The appearance and smell of the hay infusion may change week to week. This is because with time, microorganisms such as protists and bacteria will reproduce. Some are capable of decomposition, and this can cause gases to be released that smell or release byproducts that cloud the water. | |||
The purpose of this experiment was to analyze and characterize bacteria from transect #3 and to determine differences in bacterial growth in the presence of regular nutrient agar and nutrient agar + tetracycline. The initial prediction, which can be accepted, was that the plates with nutrient agar only will have more diverse and higher numbers of bacteria colonies in comparison to the nutrient agar + tetracycline plates. | |||
The effect of tetracycline on the total number of bacteria is somewhat unclear. Compared to the nutrient agar plates, the 10^-5 and 10^-7 tetracycline plates had no colonies. The 10^-3 and 10^-9 colonies did have more colonies than their sister plates without tetracycline. I believe tetracycline had an effect and limited bacteria growth significantly, however, the reason why the results appear as they are might be due to cross contamination with the spreader when placing the bacteria in the plates. The spreader was not sterilized in between spreading bacteria in the regular agar plates to the tetracycline plates. This may explain the high number of bacteria observed on the lowest concentration nutrient agar + tetracycline plate. The tetracycline seemed to cause a yellow color change in the bacteria. It appeared that one species of bacteria is unaffected by tetracycline. | |||
Tetracycline antibiotics bind to the 30S ribosomal subunit and prevent t-RNA from binding, which prevents amino acids from being added to the peptide chain, and therefore protein synthesis from occurring (HACCP). Tetracyclines affect both gram-positive and gram-negative species; some microorganisms affected include Rickettsie, Chlamydia, Mycoplasma, spirochetes, and some protozoa, but not fungi or viruses (HACCP). According to the HACCP source, resistance to tetracyclines is widespread. | |||
---- | |||
'''References''' | |||
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C. | |||
"Tetracyclines in Veterinary Medicine-Overview." ''HACCP Manual''. n.d. Bio Agri Mix. (8 July 2014). <http://www.bioagrimix.com/haccp/html/tetracyclines.htm> | |||
---- | |||
---- | |||
---- | |||
'''Lab 2: Identifying Algae and Protists''' 7/2/14 | |||
'''Introduction''' | |||
The hay infusion jar from the previous lab period can be considered to be an ecosystem, composed of many microorganisms that each have different niches, or ecological roles (Bentley 2014). To identify organisms, a dichotomous key can be employed. This type of system presents series of two choices that can be used to narrow down the organism in question to a specific species (Bentley 2014). During this lab session, the dichotomous key was employed to determine what types of protists and algae were present in the hay infusion. In addition, a serial dilution was performed to prepare for the next lab session. | |||
---- | |||
'''Materials and Methods''' | |||
Before the hay infusion culture was examined, practice using the dichotomous key was obtained by identifying known organisms. After, the hay infusion culture jar was analyzed in terms of odor and appearance. Wet mounts were prepared of samples from both the niche in the top of the jar as well as at the bottom of the jar. Next, the wet mounts were examined under microscope and two organisms from the top of the jar sample and two from the bottom of the jar sample were identified. These organisms were sketched out and pertinent information was recorded. All procedures were completed per the Bio 210 lab manual. | |||
After the hay infusion culture analysis, serial dilutions were performed in preparation for the microbiology lab. A 10^-2, 10^-4, 10^-6, and 10^-8 dilution was made and they were each spread on both a nutrient agar plate and a tetracycline plate. See below diagram of the serial dilution procedure. | |||
Figure 1: Diagram of Serial Dilution Procedure | |||
[[Image:SerialDilutionGan.png]] | |||
Serial dilution to inoculate agar petri dishes for bacterial analysis | |||
---- | |||
'''Results''' | |||
There was no strong odor coming from the jar with the hay infusion. The smell noted was similar to the odor present around most ponds. The water color observed was cloudy and a mild yellow-green tinge was noted. There was no apparent life on top of the liquid such as mold or green shoots. | |||
In the top of the jar sample, green algae and blepharisma were observed. The sample from the bottom of the jar revealed chilomonas and colpidium. Blepharisma, chilomonas, and colpidium are all protists. The algae observed was green, non-motile, and approx. 50 µm long. The blepharisma was greenish brown with a red brown pigment, non-motile, and approx. 25 µm long. The chilomonas was clear, non-motile, and 37.5 µm long. The colpidium was clear, motile, and 12.5 µm long. | |||
Figure 2: Blepharisma at 400x Magnification | |||
[[Image:Blepharisma 400x Gan.JPG]] | |||
The figure above is a photograph of blepharisma as seen through a compound microscope. | |||
---- | |||
'''Tables and Graphs''' | |||
Table 1: Organisms Observed in Hay Infusion of Transect #3 Soil | |||
[[Image:GanOrganismtablewithimages.png]] | |||
Table showing results of microscopic examination of transect hay infusion wet mounts. | |||
---- | |||
'''Discussion:''' | |||
The purpose of this lab was to observe and identify protists and algae from the transect. Samples for wet mounts were taken from the top of the jar and the bottom (two different niches) because they both have different living and nonliving factors. Organisms such as the ones identified compete for space and there is a carrying capacity, or maximum capacity for a species, in a niche (Bentley 2014). It is also possible that some organisms may be found at the top more than others due to the fact that they might need light for photosynthesis. The plant matter can help an organism by providing shelter or food, but it could also be hurtful if it blocks it from needed light. | |||
On page 2 of the Freeman textbook, the needs of life are enumerated. All organisms use energy, are made up of cells, process information, are capable of replication, and evolve (Freeman 2014). Blepharisma, a complex protist discovered in the top of the hay infusion, obtains energy by using its cilia to sweep food into its buccal cavity (Hanna). Blepharisma are unicellular, and store their information in a micronuclei, and can reproduce by binary fission or conjugation (Hanna). Conjugation confers genetic variation through recombination which can permit the organism to evolve (Freeman 2014). | |||
If the hay infusion culture had been observed for another two months, I would predict that more microbes would be visible through microscopy. I also predict that more advanced organisms would appear as the bacteria serve as the food source for other organisms, and it increases over time. There were many selective pressures that affected the community of the samples. First, they were in a space that had a set capacity. In addition, they had to survive with only the nutrients in the water as nothing was added to the jars in the couple of days of incubation. The samples had pressure from the environment, i.e. the lab temperature and light level, to survive, as the temperature and light level in the lab are different from the transect conditions. | |||
---- | |||
'''References''' | |||
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C. | |||
Freeman, Scott. 2014. Biological Science. Prentice Hall: New Jersey. 2. | |||
Hanna, Jannette. "Blepharisma". ''Micscape Magazine''. Nov. 2004. Rochester Institute of Technology. (7 July 2014). <http://www.microscopy-uk.org.uk/mag/artnov04macro/jhblepharisma.html> | |||
---- | |||
---- | |||
---- | |||
'''Lab 1: Observing a Niche at AU''' 7/1/14 | |||
'''Introduction''' | '''Introduction''' | ||
| Line 12: | Line 568: | ||
Figure 1: Map of Transect #3 Location on American University Campus | Figure 1: Map of Transect #3 Location on American University Campus | ||
[[Image:Gan Transect3 Map.jpeg]] | [[Image:Gan Transect3 Map.jpeg]] | ||
Transect #3 is located in between Bender Arena and Hughes Hall. Map from Google Maps. | Figure 1: Transect #3 is located in between Bender Arena and Hughes Hall. Map from Google Maps. | ||
---- | ---- | ||
'''Materials and Methods''' | '''Materials and Methods''' | ||
Visual analysis was completed of transect #3. The transect was walked numerous times and observations on biotic and abiotic factors were recorded. A diagram of the plants and abiotic factors was sketched out. Photos were taken from an aerial view by standing on some of the benches. In addition to the photos taken from above, some were taken at standing height to record the survey of plants, trees and bushes. | |||
Before leaving the transect, a soil and ground vegetation sample was taken. Back in the laboratory, this sample was mixed with Deer Park water and dried milk to make a hay infusion culture per lab manual instructions. The mixture was placed in an open jar and left out to be analyzed in the next lab. | |||
---- | |||
'''Results''' | '''Results''' | ||
Below is a sketch diagram of the parts of the transect and numerous photographs. The transect is on flat, level ground with the exception of some small beds that are mound shape. In addition to the trees and plants noted, some small flying insects were present in the transect. The trees create a large amount of shady area. | |||
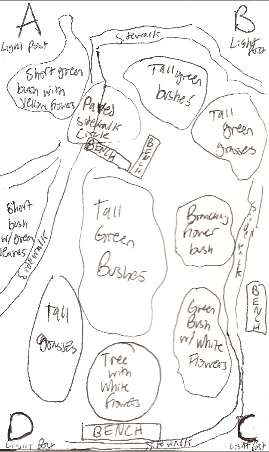
Figure 2: Diagram of Biotic and Abiotic Components of Transect #3 | |||
[[Image:Transect 3 diagram gan.png]] | |||
Transect #3 is composed of a sidewalk that loops through an area with grasses, plants, bushes and trees. | |||
Figure 3: | |||
[[Image:Picsoftranscript3 1.png]] | |||
[[Image:Picsoftranscript3 2.png]] | |||
Transect #3 hosts a variety of plant life. | |||
*Note: Aerial photography was difficult due to tall nature of transect and inability to get to a place with a higher vantage point. Instead, multiple images were taken of the variety of elements in the transect. | |||
---- | |||
'''Tables and Graphs''' | '''Tables and Graphs''' | ||
| Line 26: | Line 608: | ||
Table 1: Biotic and Abiotic Components of Transect 3 | Table 1: Biotic and Abiotic Components of Transect 3 | ||
''' | [[Image:Biotic Abiotic Gan Transect3.png]] | ||
---- | |||
'''Discussion:''' | |||
The purpose of this experiment was to explore the diversity in transcript #3. From initial observations, it appears that there are numerous types of plants and bushes present in the transect. It also appears that there are not many animals living in this transect. This may be due to the fact that the area is right next to the road and a busy part of campus that may not be conducive to undisturbed living. | |||
The results of this initial exploration will be used to further the study of the many levels of organisms in the transect. By studying a small area with biodiversity, one can learn more about ecology. | |||
---- | |||
'''References''' | '''References''' | ||
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C. | |||
Latest revision as of 13:56, 28 July 2014
TA Alyssa Pedersen MW 5:30-8:40PM
Lab 6: Analysis of Bacteria Sequencing 7/28/14
Introduction
In Lab #3, two colonies of bacteria were chosen to be set up for PCR in the hopes of having products that can be sent out for 16s sequencing. This post will present and discuss the results of Transect #3's actual bacterial sequencing results.
Bacteria are a very diverse domain, and different bacteria have their own specific enzymes (Bentley et al., 2014). The enzymes are specific due to the fact that the DNA sequences are different for the genes that code for the enzymes. PCR with a special primer that is specific for these genes can be employed to identify species, as well as show relatedness between different species (Bentley et al., 2014).
Materials and Methods
Back in Lab #3, the selected bacteria colony was transferred to a 100 microliter tube and incubated at 100 degrees C for 10 min. Next, the tube was centrifuged for 5 minutes at 13,400 rpm. A separate, labeled tube was obtained and 20mL of primer/water mixture with a PCR bead was added. 5 micro liters of the supernatant of the boiled/centrifuged samples was placed in the 16s PCR reaction, and the tube was placed in the PCR machine.
The PCR products were run on an agarose gel by the lab TAs. The PCR product was then sent out for sequencing.
The 16s sequence was analyzed using NCBI nucleotide blast to determine its identity.
All procedure explained above was followed per the 2014 Biology 210 lab manual by Bentley et al.
Results
The 16s sequence that came back from the lab was as follows: AAGATTAATACCCCATAATATTTTAAGTGGCATCACTTGAAATNGAAAACTCCGGTGGATAAAGATGGGCACGCTCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCTACGATCTTTAGGGGGCCTGANAGGGTGATCCCCCACACTGGTACTGAGACACGGACCANACTCCTACGGTAGGATCAGTGAGGAATATT
NCBI blast identified this sequence at 95% for Chryseobacterium. The corresponding accession number is JQ660730.1
From the results of the previous Bio 210 classes this year, 10 more sequences were identified using NCBI blast. These included Chryseobacterium, Flavobacterium, Pseudomonas, Flectobacillus, Sphingobacterium, and Aeromonas.
Looking back at our identifications from lab 3, the samples we identified were gram + and rods. This is different than the BLAST identified bacteria, as the ones investigated (chryseobacteria, flavobacteria, pseudomonas) are all gram - and rod shaped.
Tables and Graphs
Table 1: Bacteria Identified in Transect #3
The table above shows a sample of bacterium identified from transect #3 using NCBI Blast. The sequences identified came from both our group's sample as well as samples from previous BIO 210 classes. Once identified, the above characteristics of the bacterium were determined (Im, 2011; Genoscope, 2012; Todar, 2012). In the 11 sequences analyzed, 5 were Chryseobacteria.
Discussion:
The purpose of this lab was to identify the bacteria present in transect #3. Initally, our group discussed that we would expect a large variety of bacteria, as bacteria is so abundant and diverse. I was somewhat surprised to see that Chryseobacterium was so common and represented a large number of the sequence results. Chryseobacterium is abundant in soil, which our transect is largely composed of. Chryseobacterium, Flavobacterium, and Pseudomonas are all also abundant in water, so it is possible it has ended up in our non-aquatic transect in run-off (Im, 2011; Genoscope, 2012; Todar, 2012).
One important limitation is that the samples provided to us from previous classes were from different seasons. Our samples were from the summer. Different organisms may be present in different seasonal conditions.
The difference in our visually identified bacteria from lab #3 and the BLAST results may be due to lack of experience. Our group judged bacteria with any type of stain as gram +; we now know that the difference between gram + and - is not the presence of stain but rather the color (purple vs. pink).
References
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C.
"Flavobacterium" Genoscope. 2012. Centre National de Sequencage. (25 Jul 2014). <http://www.genoscope.cns.fr/spip/-Flavobacterium,511-.html>
Im, WT et. al. 2011. Chryseobacterium ginsenosidimutans sp. nov., a bacterium with ginsenoside-converting activity isolated from soil of a Rhus vernicifera-cultivated field. Int J Syst Evol Microbiol. 61: 1430-5
Todar, Kenneth. 2012. Todar's Online Textbook of Bacteriology. University of Wisconsin: Madison, WI.
Lab 5: Analysis of Invertebrates and Vertebrates in Transect #3 7/14/14
Introduction
In this lab, the invertebrates collected from transect #3 were analyzed. There are 14 different phyla of invertebrates, all ranging in size, shape, and complexity (Bentley 2014). Porifera, or sponges, are among the simplest invertebrates, while Platyhelminthes are more complex (Bentley 2014). Most of the invertebrate findings from transect #3 are arthropods, which consist of adult insects and their larvae (Bentley 2014). Invertebrates vary in their symmetry, embryonic tissue layers, and digestive development (Bentley 2014). The invertebrates collected from transect #3 are diverse and exemplify the variety of the invertebrate group.
Materials and Methods
All procedures were performed using the Bio 210 Lab Manual.
First, the prepared slides and samples were observed under the microscope and through the naked eye to gain experience in identifying, analyzing, and classifying invertebrates. The nematoda sample was observed for structure and movement. Some of the worms and samples arrived dead, so they were not observed.
Next, the Berlese setup was broken down to obtain the invertebrates that should have collected in the ethanol. The collection tube was poured into petri dishes for analysis, but no invertebrates were found. As none were found, the funnel leaf litter contents were dumped into a large tray. Three invertebrates were found alive and placed in the ethanol collecting tube to be analyzed with microscopy. Two samples were borrowed from other transect groups (transects #1 and #5) as our group only found 3 organisms.
Each sample was placed in a depression slide. Both a dissecting microscope and compound microscope were used to observe sample. The samples were measured and a description of the organism was recorded in the lab notebook. The dichotomous key distributed in lab as well as Google image searches were used to identify the organisms.
The last part of the lab was research of vertebrates using the textbook and internet sources. From this, a food web was created.
Results
The only worm movement examined was of the nematoda because the other samples were dead. Many nematodes were visible on the slide and there was variation in their size to some degree. The nematoda wiggled longitudinally. This fits their long slender bodies that have limited muscles to produce an undulating movement.
Figure 1: Nematoda Sample Slide
Figure 1 shows nematoda as visualized through a compound microscope.
In transect #3, our group identified a biting louse, a spider of unknown type and a armadillidiidae (pill bug). Below are pictures and descriptions of each organism. The data is organized into table 1 in the "Tables and Graphs" section.
Figure 2: Organism #1: Biting Lice
One was found in transect #3. It had six legs, antennae, hairy body, hairy legs, and was 1.125mm.
Figure 3: Organism #2: Spider of Unknown Type
One of these spiders was found in transect #3 leaf litter. The organism appeared hairy, with a large round thorax, brown in color, with 8 legs. It measured 0.625mm.
Figure 4: Organism #3: armadillidiidae (pill bug)
One was found in transect #3 leaf litter. It had about 10 legs, a segmented exoskeleton, and antennae. The sample was too large to measure via ocular micrometer.
Figure 5: Organism #4: Biting Lice
This sample came from transect #5. It had six legs, hairy antennae, and a head with a long striped body. It measured 1.95mm.
Figure 6: Organism #5: Millipede
This sample came from transect #1. It had a segmented hairy body with many legs. It measured 3.5mm.
Tables and Graphs
Table 1: Invertebrate Organisms found in Transect #3 Leaf Litter
Above are the organisms found in the transect #3 leaf litter obtained on 7/7/14. Note that the last two rows are of organisms from different transects.
Table 2: Expected Vertebrates of Transect #3
The above organisms are a prediction. The only vertebrate that has been noticed in the transect are squirrels.
Discussion:
The purpose of this lab was to identify and study different phyla of invertebrates in transect #3.
The size range is from 0.625 mm (the spider) to larger than the ocular micrometer (Armadillidiidae). It is impossible to determine exactly what kinds of organisms are most common in leaf litter because we only obtained one of each sample. From the phyla, it appears that arthropoda are common in leaf litter. Although we identified many arthropoda, there are other invertebrates that are vital to the transect.
The louse from transect #3 appeared different than the louse we borrowed from transect #5. While they both had 6 legs and hairy bodies, the louse from transect #5 had a different coloration and stripe pattern. The dichotomous key identified both organisms as biting lice, but that is just a broad category. There are over 3000 known species of lice, so it is most likely the case that each organism is a different species (Christensen 2005).
It is possible that no invertebrates were observed in the ethanol collecting tube because our leaf litter sample had too much soil in it. The thicker sample may have prevented the passage of organisms down the funnel, through the screen material, and into the collecting tube.
The identification of the sample organisms may be incorrect due to limited experience identifying invertebrates as well as the fact that the dichotomous key had a limited number of species it led to.
Five vertebrates that may inhabit the transect are the Eastern Gray Squirrel, the Eastern Garter Snake, Raccoon, the American Crow, and the American Robin. (Metropolitan Naturalist). The squirrel was observed in the transect. The four other species were predicted inhabitants after reviewing web sources on the animals native to the D.C. area. The scientific names and classification of each vertebrate show there are mammals, reptiles, and aves, but that they differ from order down, with exception to the two birds that both share the order Passeriformes. The phylum through species categorization of each vertebrate is located in Table 2.
Biotic and abiotic characteristics of the transect can benefit each species. Squirrels live in leafy nests on tree branches, and transect #3 has two large trees for squirrels to make these nests (Metropolitan Naturalist). Garter snakes eat worms and even mice, which are most likely present in the transect (Metropolitan Naturalist). Raccoons should be visitors of the transect due to the proximity of garbage cans, as they eat trash, as well as due to the smaller animals in the transect which they prey on (Metropolitan Naturalist). The transect provides tall trees for crows to perch atop, and since crows eat trash, there is trash littered by humans in the transect as people walk on the path through it. The transect can support robins because it has plenty of branches to make nests and the grass/dead leaves create a good camouflage environment for them.
The following food web has been constructed to show the relationships between transect #3 organisms.
Figure 7: Transect #3 Food Web
Food web shows energy flow between bacteria, plant matter, invertebrates, and vertebrates. This diagram is just of the identified organisms of transect #3; our group acknowledges that there are many, many more organisms that we did not identify and are not on this web.
The concept of community states describes when within a particular area, populations of different species interact (Freeman 2014). The food web shows the different organisms that live in the transect together that interact through consuming each other. Carrying capacity is the maximum population that an environment can sustain (Freeman 2014). The resources in transect #3 are limited in terms of food and housing; this affects the number of each species and the balance of organisms in the transect. Trophic levels refer to the feeding levels in an ecosystem (Freeman 2014). The food web shows the different tropic levels of transect #3, with the dead animals and leaves as well as live leaves and flowers in the first trophic level, all the way to the vertebrates, which are in the highest trophic level.
-
References
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C.
Christensen, Karin. "External Parasites". The Biology of the Goat. 2005. (16 July 2014). <http://www.goatbiology.com/lice.html>
"Some Animals of the Metro Area". Metropolitan Naturalist. (16 July 2014). <http://metro-naturalist.com/Animals/Fauna_Menu.html>
Lab 4: Studying Plants and Fungi in Transect #3 7/9/14
Introduction
Today's land plants are thought to have evolved from bryophytes, non-vascular mosses (Bentley 2014). These mosses and simple plants evolved when water space became competitive and land space was wide open. Plants evolved to tolerate the temperature differences on land, water and nutrient access issues, as well as male and female gametic interaction (Bentley 2014). Tracheophytes diverged from the bryophytes in the Devonian period and radiation occurred; in the more recent Cretaceous period, Angiosperms (flowering plants) radiated into many niches (Bentley 2014).
In this lab session, the plants and fungi on transect #3 were analyzed. Special note was taken of the vascularization, specialized structures, and reproduction anatomy of these plants.
Materials and Methods
The first procedure performed was collection of plant samples from the transect. A leaf litter sample consisting soil and dead leaves was collected into a large Ziploc bag to be used in the Berlese funnel. Next, a sample was taken from five plants in the transect in the form of a flower or leaf.
Back in the lab, we first practiced analyzing known plant and fungi samples with both the naked eye as well as through compound and dissecting microscopy. Seeds were dissected to identify the different parts. After practicing, we analyzed the five plant transect samples and recorded notes on vascularization, specialized structures, and reproduction.
The last procedure was setting up the Berlese funnel for invertebrate collection. A funnel with a screen at the base was connected to a flask filled with preservative. A light source was placed above the funnel. The soil and leaf litter sample was placed in the funnel. (Bentley 2014).
Results
No seeds were obtained from the transect.
The leaf litter sample was obtained from the main bed of the transect around the front tree and low bushes. See picture below of leaf litter collection site.
Figure 1: Leaf Litter Sample Collection Site
The leaf litter sample for the Berlese funnel was collected next to a bush and a tree and consisted of dry leaves and dirt.
The plant samples were obtained from a wide variety of plant life in the transect. The updated transect map below shows the sample number and location it was collected from. A leaf or flower was picked from each plant as a representative sample. All of the plants had vascular structures. Our group did not obtain any seeds from the transect, so we cannot be sure about plant vascularization.
Sample #1 and #5 are of angiosperms. Angiosperms are flowering plants that are a group of seed plants (Freeman 2014) . Sample #1 was a white, conical flower that was approx. 6cm in diameter. This flower came from a tall green bush. Sample #5 was a yellow, five-petal flower that was approx. 5 cm in diameter and came from a short green bush.
Table #1 in the "Tables and Graphs" section shows the characteristics of all five transect plant samples.
Figure 2: Map of Transect #3 with Plant Sample Locations
The figure above is an updated map of the transect with the numbers 1-5 to denote the locations the plant samples were taken.
Figure 3: Plant Sample #1
This plant is located near light post C; it is a green bush with white flowers.
Figure 4: Plant Sample #2
This plant is located in the front of the transect near the bench; it is a tree with white flowers.
Figure 5: Plant Sample #3
This plant is located in the middle of the transect; it is a maple tree.
Figure 6: Plant Sample #4
This plant is located in the middle of the transect; it is a tall green bush.
Figure 7: Plant Sample #5
This plant is located near light post A; it is a short green bush with yellow flowers.
Figure 8: Flowers and Leaf Samples from Plant Samples
The picture above depicts the five samples obtained. Starting from the left, they are ordered 1-5.
Figure 9: Fungi observation
The fungi observed was of Rhizopus stolonifer, commonly known as black bread mold.
Figure 10: Sketch of Fungi Under Microscope
The above sketch is of Rhizopus stolonifer, commonly known as black bread mold.
Tables and Graphs
Table 1: Characteristics of Transect #3 Plant Samples
Table 1 shows the characteristics of the five plant samples obtained from transect #3. Each plant was similar in that they were vascularized, but they each had different specialized structures. Classification on reproduction was estimated using the monocot vs. dicot table in the lab manual to characterized observed morphologies as no seeds were available for analysis.
Discussion:
Through this lab, we sought to identify and analyze plant samples from transect #3. Bushes and trees are vascular and seed producing. In the transect, we saw diversity in the leaf type and size, as well as in the flowers. The leaves varied in shape from oval to multi-pronged and they were of different shades. Some had more of a wax covering than others, and some had spiked ends, presumably for defense and protection. The flowers varied from conical shaped to petaled, and from white to yellow.
Without seeds to examine, we had to make educated guesses on whether the plant samples were monocot or dicot. Our group acknowledged that analysis of cotyledons in seeds would have been preferable to determine whether the samples are monocot or dicot. From the lab manual table, monocots usually have long narrow leaves with parallel veins and dicots have broader leaves with a network of veins (Bentley 2014). It was also possible to base the decision on floral arrangement, as monocots usually have floral parts in multiples of three and dicots in multiples of either four or five (Bentley 2014). Sample #2 and #4 had parallel veins and a more narrow leaf structure that correlates with monocots. In contrast, sample #3 was of broad shape and had a branch like vein structure. Samples #1 and #5 both had petals in multiples of four or five, which correlates with dicot structure.
Fungi sporangia are cells that holds spores until they are mature and ready to be dispersed (New Brunswick Museum). Sporangia are important because they cane be the site of meiosis or mitosis and that they are responsible for spreading fungi. Fungi can reproduce asexually by means of sporangia (New Brunswick Museum). The samples observed under the microscope appeared to be fungi. One was a Rhizopus stolonifer (bread mold), which is a zygomycota. It appeared to be a zygomycota due to the thick walled zygospore present (Bentley 2014). There are many types of fungi, but most are filamentous and fungi have cell walls and spores (Bentley 2014). The samples observed under the microscope had these features.
References
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C.
"Fungi Reproducing Asexually By Means on Sporangia". Natural History of Fungi. N.d. New Brunswick Museum. (13 July 2014) <http://website.nbm-mnb.ca/mycologywebpages/NaturalHistoryOfFungi/AsexualSporangia.html>
Lab 3: Analysis of Bacteria in Transect #3 7/7/14
Introduction
This lab focuses on the study of the bacteria found in transect #3. There are different groups of bacteria, including Proteobacteria, Chlamydiae, Actinobacteria, Spirochetes Cyanobacteria, and Firmicutes (Bentley 2014). Biologists classify bacteria on morphology, and examine characteristics such as form, elevation, margin, size, and stain characteristics (Bentley 2014). A positive gram stain yields bacteria that remains violet after preparation due to the thick peptidoglycan wall, while negative gram stains do not hold the dye (Bentley 2014).
In this lab, the agar plates prepared by serial dilution in lab 2 were analyzed by regular observation with microscopy as well as with gram staining. At the end of lab, PCR was set up for 16s sequencing to be run through gel next lab class.
I do not believe any Archaea species will have grown on the agar plates. Since Archaea tend to grow in extreme environments, the petri dish with agar will be no match. The room temperature petri dish is much cooler than a hot spring, one environment in which Archaea thrive (Bentley 2014).
I predict that the plates with nutrient agar only will have more diverse and higher numbers of bacteria colonies in comparison to the nutrient agar + tetracycline plates. If tetracycline is effective against the transect 3 bacteria, then there will be fewer colonies observed on the corresponding agar plates.
Materials and Methods
First, all six petri dishes were placed on the bench for visual inspection with the naked eye. From this, it was possible to gather general information about the number of colonies, pattern, colors, and shapes.
Next, wet mounts were prepared from two of the nutrient agar petri dishes and one of the tetracycline petri dishes. These wet mounts were examined under the compound microscope at 1000x using oil.
After, gram stain slides were prepared from the same colonies used for the three regular wet mounts. The colony samples were heat fixed to the slide, stained with crystal violet, and set with Gram's iodine mordant and safranin stain. Observations were recorded under 400x and 1000x.
The final procedure was the PCR setup for 16s sequencing. Bacteria from each of the three colonies used in the wet mount and gram stain was incubated and then centrifuged. Samples from these three tubes were then mixed with a PCR bead and placed into a smaller tube with 16s PCR reaction liquid (Bentley 2014).
Results
Our group briefly glanced at the Hay Infusion and there was nothing to report on any changes in appearance. The Hay Infusion was not opened so no observations were noted on changes in smell.
There were differences noted between the plates with antibiotic and the plates without antibiotic. All of the tetracycline plates had yellow colonies. In contrast, the nutrient agar plates varied in color, from red to white to yellow colonies. In general, there was more variation noted on the nutrient agar plates, including different edges on the colonies and the presence of a small lawn on the 10^-5 plate, as well as more colonies. The tetracycline plate colonies appear to have entirely even margins.
Our transect group chose to take samples of colonies from the 10^-9 agar+tet, 10^-9 agar, and 10^-5 agar dishes. See figures 3, 4 and 5 for images of these selected colonies. From these samples, no bacteria was motile. The 10^-9 tetracycline + agar and 10^-5 agar samples appeared gram positive, while the 10^-9 agar was gram negative. In regards to bacterial shape, the 10^-5 agar sample appeared to be coccus shaped while the 10^-9 tetracycline + agar sample was bacillus shaped. See images and tables below.
Figure 1: 100 fold Dilutions Petri Dishes (First Four)
These were grown with nutrient agar. Bacteria source is transect 3 hay infusion culture.
Figure 2: 100 fold Dilutions Petri Dishes (Second Four)
These petri dishes contained nutrient agar and tetracycline. Bacteria source is transect 3 hay infusion culture.
Figure 3: 10^-5 Nutrient Agar Petri Dish
This dish was chosen for microscopy, gram staining, and PCR. The sample was taken from one of the larger yellow colonies.
Figure 4: 10^-9 Nutrient Agar Petri Dish
This dish was chosen for microscopy, gram staining, and PCR. The sample was taken from the large red colony.
Figure 5: 10^-9 Nutrient Agar + Tetracycline Petri Dish
This dish was chosen for microscopy, gram staining, and PCR. The sample was taken from a larger yellow colony.
Figure 6: View of Gram Stained Sample from 10^-9 Tet Petri Dish
This image shows the gram positive colony of bacteria from the 10^-9 agar and tetracycline petri dish at 1000x.
Figure 7: View of Gram Stained Sample from 10^-9 Tet Petri Dish
This image shows the gram negative colony of bacteria from the 10^-9 nutrient agar petri dish at 1000x. The red seen in the photo is from the wax pencil, there is no stained bacteria present.
Figure 8: View of Gram Stained Sample from 10^-5 Petri Dish
This image shows the gram positive colony of bacteria from the 10^-5 nutrient agar petri dish at 1000x.
Tables and Graphs
Table 1: Results of Serial Dilutions in Nutrient and Nutrient + Antibiotic Agar Plates
This table shows data from colonies counted on plates with different dilutions. Nutrient agar plates had more bacterial colonies observed.
Table 2: Bacteria Characterization of 10^-9 Tet, 10^-9, and 10^-5 Plate Colonies
The data presented above is from the analysis of gram staining the three colonies selected from 10^-9 agar and tetracycline, 10^-9 nutrient agar, and 10^-5 nutrient agar.
Discussion:
The appearance and smell of the hay infusion may change week to week. This is because with time, microorganisms such as protists and bacteria will reproduce. Some are capable of decomposition, and this can cause gases to be released that smell or release byproducts that cloud the water.
The purpose of this experiment was to analyze and characterize bacteria from transect #3 and to determine differences in bacterial growth in the presence of regular nutrient agar and nutrient agar + tetracycline. The initial prediction, which can be accepted, was that the plates with nutrient agar only will have more diverse and higher numbers of bacteria colonies in comparison to the nutrient agar + tetracycline plates.
The effect of tetracycline on the total number of bacteria is somewhat unclear. Compared to the nutrient agar plates, the 10^-5 and 10^-7 tetracycline plates had no colonies. The 10^-3 and 10^-9 colonies did have more colonies than their sister plates without tetracycline. I believe tetracycline had an effect and limited bacteria growth significantly, however, the reason why the results appear as they are might be due to cross contamination with the spreader when placing the bacteria in the plates. The spreader was not sterilized in between spreading bacteria in the regular agar plates to the tetracycline plates. This may explain the high number of bacteria observed on the lowest concentration nutrient agar + tetracycline plate. The tetracycline seemed to cause a yellow color change in the bacteria. It appeared that one species of bacteria is unaffected by tetracycline.
Tetracycline antibiotics bind to the 30S ribosomal subunit and prevent t-RNA from binding, which prevents amino acids from being added to the peptide chain, and therefore protein synthesis from occurring (HACCP). Tetracyclines affect both gram-positive and gram-negative species; some microorganisms affected include Rickettsie, Chlamydia, Mycoplasma, spirochetes, and some protozoa, but not fungi or viruses (HACCP). According to the HACCP source, resistance to tetracyclines is widespread.
References
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C.
"Tetracyclines in Veterinary Medicine-Overview." HACCP Manual. n.d. Bio Agri Mix. (8 July 2014). <http://www.bioagrimix.com/haccp/html/tetracyclines.htm>
Lab 2: Identifying Algae and Protists 7/2/14
Introduction
The hay infusion jar from the previous lab period can be considered to be an ecosystem, composed of many microorganisms that each have different niches, or ecological roles (Bentley 2014). To identify organisms, a dichotomous key can be employed. This type of system presents series of two choices that can be used to narrow down the organism in question to a specific species (Bentley 2014). During this lab session, the dichotomous key was employed to determine what types of protists and algae were present in the hay infusion. In addition, a serial dilution was performed to prepare for the next lab session.
Materials and Methods
Before the hay infusion culture was examined, practice using the dichotomous key was obtained by identifying known organisms. After, the hay infusion culture jar was analyzed in terms of odor and appearance. Wet mounts were prepared of samples from both the niche in the top of the jar as well as at the bottom of the jar. Next, the wet mounts were examined under microscope and two organisms from the top of the jar sample and two from the bottom of the jar sample were identified. These organisms were sketched out and pertinent information was recorded. All procedures were completed per the Bio 210 lab manual.
After the hay infusion culture analysis, serial dilutions were performed in preparation for the microbiology lab. A 10^-2, 10^-4, 10^-6, and 10^-8 dilution was made and they were each spread on both a nutrient agar plate and a tetracycline plate. See below diagram of the serial dilution procedure.
Figure 1: Diagram of Serial Dilution Procedure
Serial dilution to inoculate agar petri dishes for bacterial analysis
Results
There was no strong odor coming from the jar with the hay infusion. The smell noted was similar to the odor present around most ponds. The water color observed was cloudy and a mild yellow-green tinge was noted. There was no apparent life on top of the liquid such as mold or green shoots.
In the top of the jar sample, green algae and blepharisma were observed. The sample from the bottom of the jar revealed chilomonas and colpidium. Blepharisma, chilomonas, and colpidium are all protists. The algae observed was green, non-motile, and approx. 50 µm long. The blepharisma was greenish brown with a red brown pigment, non-motile, and approx. 25 µm long. The chilomonas was clear, non-motile, and 37.5 µm long. The colpidium was clear, motile, and 12.5 µm long.
Figure 2: Blepharisma at 400x Magnification
The figure above is a photograph of blepharisma as seen through a compound microscope.
Tables and Graphs
Table 1: Organisms Observed in Hay Infusion of Transect #3 Soil
Table showing results of microscopic examination of transect hay infusion wet mounts.
Discussion: The purpose of this lab was to observe and identify protists and algae from the transect. Samples for wet mounts were taken from the top of the jar and the bottom (two different niches) because they both have different living and nonliving factors. Organisms such as the ones identified compete for space and there is a carrying capacity, or maximum capacity for a species, in a niche (Bentley 2014). It is also possible that some organisms may be found at the top more than others due to the fact that they might need light for photosynthesis. The plant matter can help an organism by providing shelter or food, but it could also be hurtful if it blocks it from needed light.
On page 2 of the Freeman textbook, the needs of life are enumerated. All organisms use energy, are made up of cells, process information, are capable of replication, and evolve (Freeman 2014). Blepharisma, a complex protist discovered in the top of the hay infusion, obtains energy by using its cilia to sweep food into its buccal cavity (Hanna). Blepharisma are unicellular, and store their information in a micronuclei, and can reproduce by binary fission or conjugation (Hanna). Conjugation confers genetic variation through recombination which can permit the organism to evolve (Freeman 2014).
If the hay infusion culture had been observed for another two months, I would predict that more microbes would be visible through microscopy. I also predict that more advanced organisms would appear as the bacteria serve as the food source for other organisms, and it increases over time. There were many selective pressures that affected the community of the samples. First, they were in a space that had a set capacity. In addition, they had to survive with only the nutrients in the water as nothing was added to the jars in the couple of days of incubation. The samples had pressure from the environment, i.e. the lab temperature and light level, to survive, as the temperature and light level in the lab are different from the transect conditions.
References
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C.
Freeman, Scott. 2014. Biological Science. Prentice Hall: New Jersey. 2.
Hanna, Jannette. "Blepharisma". Micscape Magazine. Nov. 2004. Rochester Institute of Technology. (7 July 2014). <http://www.microscopy-uk.org.uk/mag/artnov04macro/jhblepharisma.html>
Lab 1: Observing a Niche at AU 7/1/14
Introduction
The American University campus is saturated with different varieties of ecosystems. This lab notebook entry, the first in a series of three, will present and discuss observations from transect #3. Through this transect, it will be possible to study the biology of communities. The biodiversity, or number and type of species that live in an area, will be analyzed (Bentley 2014). In addition, living (biotic) and nonliving (abiotic) elements will be examined (Bentley 2014). Each species in the community has a niche that is important to the status of the community.
Transect #3 is located between Bender Arena, Hughes Hall, and the Woods-Brown Amphitheater. The coordinates are 38°56'16.2"N 77°05'20.4"W. The transect, approximately 20mx20m, has a collection of bushes, grasses, plants, and trees.
Figure 1: Map of Transect #3 Location on American University Campus
Figure 1: Transect #3 is located in between Bender Arena and Hughes Hall. Map from Google Maps.
Materials and Methods
Visual analysis was completed of transect #3. The transect was walked numerous times and observations on biotic and abiotic factors were recorded. A diagram of the plants and abiotic factors was sketched out. Photos were taken from an aerial view by standing on some of the benches. In addition to the photos taken from above, some were taken at standing height to record the survey of plants, trees and bushes.
Before leaving the transect, a soil and ground vegetation sample was taken. Back in the laboratory, this sample was mixed with Deer Park water and dried milk to make a hay infusion culture per lab manual instructions. The mixture was placed in an open jar and left out to be analyzed in the next lab.
Results
Below is a sketch diagram of the parts of the transect and numerous photographs. The transect is on flat, level ground with the exception of some small beds that are mound shape. In addition to the trees and plants noted, some small flying insects were present in the transect. The trees create a large amount of shady area.
Figure 2: Diagram of Biotic and Abiotic Components of Transect #3
Transect #3 is composed of a sidewalk that loops through an area with grasses, plants, bushes and trees.
Figure 3:
Transect #3 hosts a variety of plant life.
- Note: Aerial photography was difficult due to tall nature of transect and inability to get to a place with a higher vantage point. Instead, multiple images were taken of the variety of elements in the transect.
Tables and Graphs
Table 1: Biotic and Abiotic Components of Transect 3
Discussion:
The purpose of this experiment was to explore the diversity in transcript #3. From initial observations, it appears that there are numerous types of plants and bushes present in the transect. It also appears that there are not many animals living in this transect. This may be due to the fact that the area is right next to the road and a busy part of campus that may not be conducive to undisturbed living.
The results of this initial exploration will be used to further the study of the many levels of organisms in the transect. By studying a small area with biodiversity, one can learn more about ecology.
References
Bentley, M., Walters-Conte, & Zeller, N.K. 2014. A Laboratory Manual to Accompany General Biology II. American University Department of Biology: Washington, D.C.