User:Eliza B/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| (26 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
'''3/21 EB''' | |||
'''Zebrafish Experiment''' | |||
''Introduction'' | |||
The purpose of the zebrafish experiment was to determine the effects fluoride on zebrafish development. | |||
Though zebrafish are not mammals, they share many similarities in their embryonic development to humans. | |||
Information gathered from experiments on zebrafish development can have implications on human embryonic development. | |||
They hypothesis of the experiment was that zebrafish in the NaF group would be stunted and have weak movement and slow startle responses. | |||
''Materials and Methods'' | |||
This study followed zebrafish over the course of two weeks as they developed from fertilized eggs to tadpoles. To set up the experiment, we obtained two identical petri dishes and designated one as a control dish and the second as the test dish. The control dish was filled with 20mL of filtered Deerpark water using a motorized pipette filler. The fluoride dish was filled with 20mL of a solution of .1% sodium fluoride. Two transfer pipettes were obtained, and the tips of each pipette were cut with scissors. 20 zebrafish eggs were transferred from a container to the control dish using a transfer pipette. 20 more zebrafish eggs were transferred using a second transfer pipette. The two petri dishes were then observed under a dissecting microscope to determine the life stage of the zebrafish. | |||
Despite efforts to use sterile instruments and clean water, the first batch of zebrafish became moldy after less than two days. Therefore, two days after setting up the experiment, the steps described above had to be repeated with new zebrafish eggs. Particular care was taken to choose eggs with little mold. After the experiment was successfully set up, the fish were monitored and petri dishes cleaned once every Monday, Wednesday, and Friday for two weeks. One partner came in on Mondays while the other came in on Fridays. Both partners participated in monitoring and observing the zebrafish on Wednesdays. | |||
Each day we came in, we first counted the number of zebrafish hatched, the number dead, and the number alive in both the control and test group. We observed the fish to see if there were any obvious differences in movement, size, or body shape. We also observed the fish under a dissecting microscope to note color and to find a fish representative of the group. Once a representative fish from each section was found, we used a transfer pipette to take the fish from the petri dish to a depression slide. We measured the pupil size, eye size, and body length under the 4x objective on a microscope. If possible, we attempted to take the heart rate as well. We also observed eye movement and attempted to calculate moves per minute. We chose eye and body size and movement-related measurements because previous research on the subject suggested that sodium fluoride stunted head-to-tail length and inhibited movement in developing embryos and tadpoles. | |||
After observing a fish from the control and the test group, we used a transfer pipette to bring them back to the petri dish. We cleaned the fluid in each dish, using a transfer pipette to remove any dirt, leftover food, or mold we found in the fluid and place it into a waste container. Any egg shells or dead zebrafish were also pipetted out and placed in the waste container. The dirty fluid we took out, was then replaced with 20mL of new fluid. More new fluid was added each day than taken out to make up for the effects of evaporation. Once the fish had hatched from their eggs, we also added a drop of food to each dish each day we came in. Only one drop was added to keep the leftover food from molding. After each day, we recorded our data into a Google spreadsheet. | |||
Several days into the experiment, after all the zebrafish had hatched, we selected one specimen from each group to be sacrificed for observation. We put each hatchling in a separate labeled tube. To each tube we added a tricaine solution, and the TA added paraformaldehyde. The TA then fixed the samples, which we observed in a depression slide under the microscope the next week. To end the experiment, we moved the live control fish into a large bowl with other live control fish, and we put the live test fish in a different bowl with other live test fish. We then discarded the dead fish and leftover fluid into a waste container. | |||
''Data'' | |||
[[Image:screen1.png]] | |||
[[Image:screen2.png]] | |||
[[Image:screen3.png]] | |||
[[Image:screen4.png]] | |||
[[Image:screen5.png]] | |||
''Conclusion'' | |||
The data show that the zebrafish experiment provided evidence in favor of the hypothesis in some cases and provided evidence against the hypothesis in other cases. As predicted, the test group was smaller in size, both head-to-tail length and eye size. However, unlike the predictions, the test group was faster, healthier, and more vibrant than the control group. The control group did not have strong startle responses, while the test group had frequent movement. This could be because the control group's petri dish was often much dirtier than the test groups. Perhaps the negative effects of fluoride on development were outweighed by fluoride's ability to clean the water or prevent bacterial growth. Future research should test zebrafish in containers that can be fully cleaned on a regular basis to avoid this complicating factor. | |||
'''2/18 EB''' | |||
'''Lab 5: Invertebrates''' | |||
''Introduction'' | |||
Though they are small, invertebrates make up a huge percentage of world’s biodiversity. With 14 different phyla and many species ranging from microscopic mites to bird-sized butterflies, invertebrates are an integral part of any ecosystem. This lab’s goal was to understand the diversity of invertebrates that lived within the wildlife habitat transect. | |||
''Materials and Methods'' | |||
We first destructed the Berlese Funnel and took samples from the ethanol in the tube below. Using a transfer pipette, we put a sample from the top of the tube into a petri dish and put a sample from the bottom into a petri dish. We then looked at the petri dishes under a dissecting tunnel to find any invertebrates. Once we found an invertebrate, we used a dichotomous key to identify the organism. | |||
''Data'' | |||
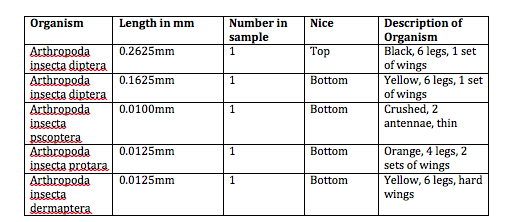
There were very few invertebrates in our petri dishes. Though we were able to find five after much searching, our transect clearly contained far fewer invertebrates than any other type of organism we have studied thus far. Every invertebrate we found was small, ranging from 0.01mm to 0.2625mm. We found two different kinds of flies, the largest organisms in the sample. The smallest organism was the psocoptera, which was a winged insect like the flies. The most common invertebrates in the wildlife habitat appear to be small winged invertebrates like flies and psocopteras. Below is a table detailing our results and some pictures of our invertebrates. | |||
[[Image:Bugtable.png]] | |||
[[Image:bug1.JPG]] | |||
[[Image:bug2.JPG]] | |||
[[Image:bug3.JPG]] | |||
There are few vertebrates that in habit our transect. Squirrels thrive in the trees, while we have seen rats scurrying on the ground. Mice might also be likely to thrive in the wildlife habitat. Birds like sparrows and robins would also be likely to make their homes in the habitat. However, since it is winter, few vertebrates are outside. Aside from squirrels and rats, we have seen no vertebrates in our transect. Below is a table of the five possible species that inhabit our transect and a list of biotic and abiotic factors that might benefit them. | |||
[[Image:Vertebrate.png]] | |||
Below is a food web that shows the interdependence of species in a transect. | |||
[[Image:themchains.JPG]] | |||
All of these species live in the same ecosystem and therefore are in community with one another. Since they all rely on base food sources like insects and seeds (whether they are the hawk that eats the seed-eating animals or the seed-eating animals themselves), their size is limited by the amount of base food available to the species at the bottom of the food chain. Species like the sparrow and the robin that rely on the same type of food source occupy the same trophic level and are further limited because they are in direct competition with another species for food (Freeman 2008). Each species has a carry capacity, or the number of organisms that can remain at a stable population within the given niche. A species carrying capacity is determined by the amount of food, predators, and competitors in the area. | |||
''Conclusion'' | |||
This lab showed that few invertebrates thrived in the wildlife transect. Since many insects hibernate or migrate over the winter, it is possible that the biodiversity of the transect is larger in the spring and summer months. Despite the lack of invertebrates, however, they play a vital role in the food chain and ecosystem of the wildlife habitat, supplying food for small mammals and birds and gaining their nutrition from the plants within the habitat. The ecosystem would be incomplete without invertebrates. | |||
''References'' | |||
Freeman, Scott. 2008. Biological Science. Pearson Benjamin Cummings: San Francisco. 130. Print. | |||
'''2/10 EB''' | |||
'''Lab 4: Plants and Fungi''' | |||
''Introduction'' | |||
As one of the core providers of oxygen to the atmosphere and a vital food source for humans and wildlife alike, plants are an irreplaceable part of any ecosystem. The core goal of this lab was to identify plants within the Wildlife Habitat transect and to analyze their structures and compositions. Without knowing the kinds of plants within the transect, it is impossible to understand the biodiversity of the ecosystem as a whole. | |||
''Materials and Methods'' | |||
This lab was divided into three parts. First we went to the Wildlife Habitat Transect and gather leaf samples from five different plants as well as 500g of leaf litter from the ground. We then examined several known samples of plants under the microscope to get a grasp on the different kinds of plants and fungi. Once we felt confident in our abilities, we cut cross sections from each of the five plants, made a wet mount from the cross sections, and analyzed each we mount under a microscope. We noted the size of the cells, and vascularization, and any specialized cells. The next step was to examine different prepared slides of fungi and identify which of the three fungi groups they belonged to. The third step in lab was to create a Berlese Funnel to collect invertebrates. To do this, we took a small plastic tube and filled it with 25mL of ethanol. We taped a funnel to this tube and laid a piece of black mesh over the funnel’s hole. We then stuffed the funnel full of our leaf litter samples and hooked the entire contraption to a pole underneath a light source. The Berlese Funnel will catch invertebrates in the ethanol below, while the black mesh will keep the leaf litter from contaminating the ethanol. The entire class’s Berlese Funnels were then covered in aluminum foil to keep the light locked in. | |||
''Data and Observations'' | |||
Since it was wintertime, there were few plants alive in our transect. We took a leaf from a medium sized tree in the bottom right corner of the transect whose leaves grew in patterns of three from a central point. We took another sample from a small tree in the top right that had only dead leaves. This tree’s leaves seemed to only grow one at a time, each from a different starting point. We gathered samples from two different kinds of monkey grass that grew near the rock path in the middle of the transect. One type had very long, thin, light green leaves, and the other had shorter, thicker, darker green leaves. Both grasses had leaves that originated from one center in the ground. Our last sample was from a purple plant that had layers of leaves growing up a central stalk. This plant came from the bottom left corner of the transect. Though it was difficult to tell in the winter when no evidence of flowering exists, we belief most of our plants were angiosperms, or flowering plants. The purple plant still had evidence of a flowering body on its head and grasses are known to be angiosperm. The two trees were harde''Italic text''r to tell, but since the smaller tree was clearly seasonal, it is safe to assume it is an angiosperm. The live tree, however, was harder to tell. Since it had visible veins and broad leaves, we again assumed it was an angiosperm. Below is a map with the locations of the plants circled. | |||
[[Image:12345.JPG]] | |||
Below are pictures of the five different plants in the transect and a close up picture of the sample we took from each plant. All microscopic pictures were taken at 10x. | |||
1: Monkey Grass 1 | |||
[[Image:monkey.JPG]] | |||
[[Image:monkleaf.JPG]] | |||
[[Image:monkcell.JPG]] | |||
2: Green Tree | |||
[[Image:Live.JPG]] | |||
[[Image:liveleaf.JPG]] | |||
[[Image:live1.jpeg]] | |||
3: Monkey Grass 2 | |||
[[Image:monkeysm.JPG]] | |||
[[Image:smallleaf.JPG]] | |||
[[Image:grass1.jpg]] | |||
4: Purple Plant | |||
[[Image:Purple.JPG]] | |||
[[Image:purpleaf.JPG]] | |||
[[Image:purpcell.JPG]] | |||
5: Dead Tree | |||
[[Image:Dead.JPG]] | |||
[[Image:deadleaf.JPG]] | |||
[[Image:red1.jpg]] | |||
When we examined the cross sections under the microscope, we noticed that the monkey grass had the clearest vascularization. The light green monkey grass had wide, vertical veins evident on the cellular level, and the darker green grass had smaller veins. The purple plant was multicolored on a cellular level, with each cell a different shade of purple, red, orange, or green. Below is a table of our findings. | |||
[[Image:shot1.png]] | |||
We then examined fungi. I looked at agaricus bisporus, rhizopus stoloniger zygospore, and rhizopus nigricons sporangia. Agaricus bisporus, or the common mushroom, is a basidiomycota gunfi because of its special gill-like structures on its underside that allow for sexual reproduction. Rhizopus stolonifer zygospore, or black bread mold, is a zygomycota because it reproduces by creating a zygospore. These fungi structures called sporangia that contain spores. These spores are the way that zygomycota fungi reproduce and are also the reason that black bread mold has its distinct black color. Finally, rhizopus nigricans sporangia is also a zygomycota. Below is a drawing and a picture of rhizopus nigricans sporangia. The bulb-like structure shown is the sporangia. | |||
[[Image:111.JPG]] | |||
[[Image:spore.JPG]] | |||
''Conclusions and Further Study'' | |||
The diversity of plants is astounding. After analyzing only five plants from the Wildlife Habitat transect, the differences in size, structure, and vascularization were plain. Each plant provides different resources to its ecosystem and requires different conditions to survive. This lab provided a greater understanding of the diversity of the world of plants and their impacts on an ecosystem. | |||
'''2/2 EB''' | |||
'''Lab 3: Bacteria in Transect 2''' | |||
''Introduction'' | |||
The core goal of this lab was to observe the bacteria that grew in transect 2 and to understand antibiotic resistance in bacteria. After plating serial dilutions of samples from our Hay Infusion onto both nutrient and antibiotic-laced plates, my group hypothesized that the plates with antibiotics on them will have less bacteria growing on them. This is intuitive because bacteria will have a harder time growing in environments with particles designed to destroy them. We also predict that there will be no Archaea species in our plates. Archaea typically live in very harsh environments, and the wildlife habitat of transect two is not harsh at all. Therefore, though this lab described different Archaea species, we do not anticipate finding any of them in our samples. | |||
''Materials and Methods'' | |||
First we observed our Hay Infusions to see how they had changed over the course of the week. After taking several pictures of the infusion, we moved on to analyzing the bacteria plates. We collected our plates and observed the color and size of both the nutrient and the tetracycline (henceforth referred to as tet) plates. We counted, as best we could, the number of colonies on each plate and recorded the information. We next created two wet slides of the bacteria. We took one sample from the clear bacteria growing in the 10-9 nutrient plate and one sample from the orange bacteria growing in the 10-3 tet plate. Using a metal loop, we put the samples on a slide and observed them as best we could. Without staining, however, it was difficult to find and see the bacteria clearly. The next step, then, was to stain the bacteria. We did this by taking four chosen slides (10-3 and 10-9 in both nutrient and tet plates) and sealing the bacteria into place by passing it several times over a flame. After the bacteria was heat fixed, we put the slides over a dying bin and washing them with crystal violet die, iodine mordant, alcohol, and safranin stain. Between each stain, we washed the entire slide with water to clear the previous stain away. Once the bacteria was stained, washed, and dried, we used microscopes to exam each sample and determine whether it was gram positive or negative. If the color remained in the bacteria it was gram positive, and if there was no color, it was gram negative. Once the observation was complete, we took four samples from the agar plates again to prepare for PCR. We transferred the bacteria into 100 microliters of water, incubated it at 100°C for 10 minutes, and centrifuged it for 5 minutes. We then added water to a PCR tube to dissolve the bead and poured the supernatant from the centrifuged tubes into the PCR tube. We then placed it in the PCR machine. | |||
''Data and Observations'' | |||
The Hay Infusion appeared to have settled into two distinct groups. The heavier materials had fallen to the bottom and formed an ecosystem of their own, while the water throughout the rest of the jar appeared much less dense. The Hay Infusion had increased drastically in smelliness, stinking of rotting animals. Below are pictures of the Hay Infusion. | |||
[[Image:Jartop.JPG]] | |||
[[Image:Jargross.JPG]] | |||
[[Image:Jarside.JPG]] | |||
Below is a picture of the data we recorded from our agar plates. The nutrient agar plates appeared to have two different kinds of bacteria. The first was bright orange, shiny, and abundant in both the tet and the nutrient plates. The second was clear, smaller, and much harder to see. Though the tet and nutrient plates had similar numbers of the orange bacteria in each, the clear bacteria only showed up in the nutrient plate. This suggests that the clear bacteria was not resistant to tetracyclinee, but that the orange bacteria was almost completely resistant, since it showed no decreases in number between the two plates. This provides support for our hypothesis that the nutrient plates would have more bacteria in them. | |||
Tetracycline is an antibiotic that works as a protein inhibitor. It blocks tRNA from attaching to the A site in protein synthesis (Chopra and Roberts 2001). It is particularly potent to bacterial cells because they pump tetracycline into their cytoplasms, while many eukaryotic cells do not. Tetracycline more easily navigates the cell walls of gram-negative bacteria. Though originally the antibiotic was affective on many bacteria, gram positive or negative, exposure to the antibiotic has allowed many organisms, especially the gram positive, to become resistant (Chopra and Roberts 2001). | |||
Table 1 | |||
[[Image:1Table1.JPG]] | |||
10-3 Nutrient Plate | |||
[[Image:tray3.JPG]] | |||
10-5 Nutrient Plate | |||
[[Image:Tray5.JPG]] | |||
10-7 Nutrient Plate | |||
[[Image:Tray7.JPG]] | |||
10-9 Nutrient Plate | |||
[[Image:Tray9.JPG]] | |||
10-3 Tet Plate | |||
[[Image:3tet.JPG]] | |||
10-5 Tet Plate | |||
[[Image:5tet.JPG]] | |||
10-7 Tet Plate | |||
[[Image:7tet.JPG]] | |||
10-9 Tet Plate | |||
[[Image:9tet.JPG]] | |||
The wet mount samples were difficult to find since they weren’t stained. The sample of the clear bacteria was much larger and easier to see than the sample of the orange bacteria, shown as the fuzzy white dot below. None of the cells seemed motile. | |||
Wet Mount of 10-3 Tet Orange Bacteria | |||
[[Image:3nwm.JPG]] | |||
Wet Mount of 10-9 Nutrient Clear Bacteria | |||
[[Image:9ncwet.JPG]] | |||
Below are pictures and a chart of the results of our gram staining. | |||
Table 2 | |||
[[Image:Table2.JPG]] | |||
10-3 Nutrient Orange Bacteria (40x) | |||
[[Image:3nc.JPG]] | |||
10-9 Nutrient Clear Bacteria (40x) - Note the multiple different shapes the bacteria takes on. | |||
[[Image:9nc.JPG]] | |||
10-3 Tet Orange Bacteria (40x) | |||
[[Image:3tc.JPG]] | |||
10-9 Tet Bacteria (40x) - There are two pictures of the -9 tet bacteria to show the differences in appearance. | |||
[[Image:9tc.JPG]] | |||
[[Image:9tc2.JPG]] | |||
''Conclusions and Further Study'' | |||
This lab revealed that at least two different kinds of bacteria were living in our transect. One with small and clear colonies and the other with large orange colonies, both bacteria were gram positive. However, only the clear bacteria was affected by the antibiotic. This data supports our hypothesis that fewer bacteria would thrive in the tet plates and our hypothesis that no Archaea species would live in our transect. There was a core source of error in our data, however. The staining process was messy, making it difficult to tell whether cells truly were gram positive or gram negative. Future studies should address this issue by increasing the washing and cleaning time during the staining process. | |||
''Works Cited'' | |||
Chopra, I., & Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiology and molecular biology reviews, 65(2), 232-260. | |||
'''1/25 EB''' | '''1/25 EB''' | ||
| Line 9: | Line 273: | ||
''Materials and Methods'' | ''Materials and Methods'' | ||
The core goal of this lab was to identify different protists that lived in the Hay Infusion made from a sample of our transect’s dirt. We first practiced identifying protists using a dichotomous key. Once we felt comfortable finding and identifying protists, we moved on to studying protists from our Hay infusion. We took samples using a transfer pipette from both the surface and the bottom of the Hay Infusion, and placed them in separate flat disk containers. We took a sample from approximately 1cm from the bottom in the middle of the jar, and one sample right below the growths on the surface. We tried to take samples from the growths themselves, but they were too large to be transferred by the pipette. With two lids of water representing the two different niches, we then made three wet mounts of each niche. Each member of our group took one wet mount from the top and one from the bottom to observe. We tried to find three specimens from the top and three from the bottom using the dichotomous key to identify any cells we found. For each specimen, we recorded its appearance, size, and scientific name. | The core goal of this lab was to identify different protists that lived in the Hay Infusion made from a sample of our transect’s dirt. We first practiced identifying protists using a dichotomous key. Once we felt comfortable finding and identifying protists, we moved on to studying protists from our Hay infusion. We took samples using a transfer pipette from both the surface and the bottom of the Hay Infusion, and placed them in separate flat disk containers. We took a sample from approximately 1cm from the bottom in the middle of the jar, and one sample right below the growths on the surface. We tried to take samples from the growths themselves, but they were too large to be transferred by the pipette. With two lids of water representing the two different niches, we then made three wet mounts of each niche. Each member of our group took one wet mount from the top and one from the bottom to observe. We tried to find three specimens from the top and three from the bottom using the dichotomous key to identify any cells we found. For each specimen, we recorded its appearance, size, and scientific name. At the end of the lab, we created a serial dilution to prepare for our bacteria lab the next week. We had four plates of agar and four plates of agar plus an antibiotic, tetracycline. We then took 100 microliters of our Hay Infusion (thoroughly mixed), and put it into a tube with 10mL of broth. From there, we took 100µL and added it to a different tube with broth. From the second tube we took 100µL and added it to a third, and from the third we took 100µL and added it to the fourth. This is a serial dilution. Each successive tube is more diluted than the one before it. Below is a picture detailing the methods for this dilution. | ||
[[Image:ThingforIt.JPG]] | |||
''Data and Observations'' | ''Data and Observations'' | ||
| Line 35: | Line 301: | ||
Bentley, Meg. 2015. A Laboratory Manual to Accompany: General Biology II. Department of Biology, American University. | Bentley, Meg. 2015. A Laboratory Manual to Accompany: General Biology II. Department of Biology, American University. | ||
Freeman, Scott. 2008. Biological Science. Pearson Benjamin Cummings: San Francisco. 130. Print. | Freeman, Scott. 2008. Biological Science. Pearson Benjamin Cummings: San Francisco. 130. Print. | ||
Latest revision as of 08:10, 21 March 2015
3/21 EB
Zebrafish Experiment
Introduction
The purpose of the zebrafish experiment was to determine the effects fluoride on zebrafish development.
Though zebrafish are not mammals, they share many similarities in their embryonic development to humans.
Information gathered from experiments on zebrafish development can have implications on human embryonic development.
They hypothesis of the experiment was that zebrafish in the NaF group would be stunted and have weak movement and slow startle responses.
Materials and Methods
This study followed zebrafish over the course of two weeks as they developed from fertilized eggs to tadpoles. To set up the experiment, we obtained two identical petri dishes and designated one as a control dish and the second as the test dish. The control dish was filled with 20mL of filtered Deerpark water using a motorized pipette filler. The fluoride dish was filled with 20mL of a solution of .1% sodium fluoride. Two transfer pipettes were obtained, and the tips of each pipette were cut with scissors. 20 zebrafish eggs were transferred from a container to the control dish using a transfer pipette. 20 more zebrafish eggs were transferred using a second transfer pipette. The two petri dishes were then observed under a dissecting microscope to determine the life stage of the zebrafish. Despite efforts to use sterile instruments and clean water, the first batch of zebrafish became moldy after less than two days. Therefore, two days after setting up the experiment, the steps described above had to be repeated with new zebrafish eggs. Particular care was taken to choose eggs with little mold. After the experiment was successfully set up, the fish were monitored and petri dishes cleaned once every Monday, Wednesday, and Friday for two weeks. One partner came in on Mondays while the other came in on Fridays. Both partners participated in monitoring and observing the zebrafish on Wednesdays. Each day we came in, we first counted the number of zebrafish hatched, the number dead, and the number alive in both the control and test group. We observed the fish to see if there were any obvious differences in movement, size, or body shape. We also observed the fish under a dissecting microscope to note color and to find a fish representative of the group. Once a representative fish from each section was found, we used a transfer pipette to take the fish from the petri dish to a depression slide. We measured the pupil size, eye size, and body length under the 4x objective on a microscope. If possible, we attempted to take the heart rate as well. We also observed eye movement and attempted to calculate moves per minute. We chose eye and body size and movement-related measurements because previous research on the subject suggested that sodium fluoride stunted head-to-tail length and inhibited movement in developing embryos and tadpoles. After observing a fish from the control and the test group, we used a transfer pipette to bring them back to the petri dish. We cleaned the fluid in each dish, using a transfer pipette to remove any dirt, leftover food, or mold we found in the fluid and place it into a waste container. Any egg shells or dead zebrafish were also pipetted out and placed in the waste container. The dirty fluid we took out, was then replaced with 20mL of new fluid. More new fluid was added each day than taken out to make up for the effects of evaporation. Once the fish had hatched from their eggs, we also added a drop of food to each dish each day we came in. Only one drop was added to keep the leftover food from molding. After each day, we recorded our data into a Google spreadsheet. Several days into the experiment, after all the zebrafish had hatched, we selected one specimen from each group to be sacrificed for observation. We put each hatchling in a separate labeled tube. To each tube we added a tricaine solution, and the TA added paraformaldehyde. The TA then fixed the samples, which we observed in a depression slide under the microscope the next week. To end the experiment, we moved the live control fish into a large bowl with other live control fish, and we put the live test fish in a different bowl with other live test fish. We then discarded the dead fish and leftover fluid into a waste container.
Data
Conclusion
The data show that the zebrafish experiment provided evidence in favor of the hypothesis in some cases and provided evidence against the hypothesis in other cases. As predicted, the test group was smaller in size, both head-to-tail length and eye size. However, unlike the predictions, the test group was faster, healthier, and more vibrant than the control group. The control group did not have strong startle responses, while the test group had frequent movement. This could be because the control group's petri dish was often much dirtier than the test groups. Perhaps the negative effects of fluoride on development were outweighed by fluoride's ability to clean the water or prevent bacterial growth. Future research should test zebrafish in containers that can be fully cleaned on a regular basis to avoid this complicating factor.
2/18 EB
Lab 5: Invertebrates
Introduction
Though they are small, invertebrates make up a huge percentage of world’s biodiversity. With 14 different phyla and many species ranging from microscopic mites to bird-sized butterflies, invertebrates are an integral part of any ecosystem. This lab’s goal was to understand the diversity of invertebrates that lived within the wildlife habitat transect.
Materials and Methods
We first destructed the Berlese Funnel and took samples from the ethanol in the tube below. Using a transfer pipette, we put a sample from the top of the tube into a petri dish and put a sample from the bottom into a petri dish. We then looked at the petri dishes under a dissecting tunnel to find any invertebrates. Once we found an invertebrate, we used a dichotomous key to identify the organism.
Data
There were very few invertebrates in our petri dishes. Though we were able to find five after much searching, our transect clearly contained far fewer invertebrates than any other type of organism we have studied thus far. Every invertebrate we found was small, ranging from 0.01mm to 0.2625mm. We found two different kinds of flies, the largest organisms in the sample. The smallest organism was the psocoptera, which was a winged insect like the flies. The most common invertebrates in the wildlife habitat appear to be small winged invertebrates like flies and psocopteras. Below is a table detailing our results and some pictures of our invertebrates.
There are few vertebrates that in habit our transect. Squirrels thrive in the trees, while we have seen rats scurrying on the ground. Mice might also be likely to thrive in the wildlife habitat. Birds like sparrows and robins would also be likely to make their homes in the habitat. However, since it is winter, few vertebrates are outside. Aside from squirrels and rats, we have seen no vertebrates in our transect. Below is a table of the five possible species that inhabit our transect and a list of biotic and abiotic factors that might benefit them.
Below is a food web that shows the interdependence of species in a transect.
All of these species live in the same ecosystem and therefore are in community with one another. Since they all rely on base food sources like insects and seeds (whether they are the hawk that eats the seed-eating animals or the seed-eating animals themselves), their size is limited by the amount of base food available to the species at the bottom of the food chain. Species like the sparrow and the robin that rely on the same type of food source occupy the same trophic level and are further limited because they are in direct competition with another species for food (Freeman 2008). Each species has a carry capacity, or the number of organisms that can remain at a stable population within the given niche. A species carrying capacity is determined by the amount of food, predators, and competitors in the area.
Conclusion
This lab showed that few invertebrates thrived in the wildlife transect. Since many insects hibernate or migrate over the winter, it is possible that the biodiversity of the transect is larger in the spring and summer months. Despite the lack of invertebrates, however, they play a vital role in the food chain and ecosystem of the wildlife habitat, supplying food for small mammals and birds and gaining their nutrition from the plants within the habitat. The ecosystem would be incomplete without invertebrates.
References
Freeman, Scott. 2008. Biological Science. Pearson Benjamin Cummings: San Francisco. 130. Print.
2/10 EB
Lab 4: Plants and Fungi
Introduction
As one of the core providers of oxygen to the atmosphere and a vital food source for humans and wildlife alike, plants are an irreplaceable part of any ecosystem. The core goal of this lab was to identify plants within the Wildlife Habitat transect and to analyze their structures and compositions. Without knowing the kinds of plants within the transect, it is impossible to understand the biodiversity of the ecosystem as a whole.
Materials and Methods
This lab was divided into three parts. First we went to the Wildlife Habitat Transect and gather leaf samples from five different plants as well as 500g of leaf litter from the ground. We then examined several known samples of plants under the microscope to get a grasp on the different kinds of plants and fungi. Once we felt confident in our abilities, we cut cross sections from each of the five plants, made a wet mount from the cross sections, and analyzed each we mount under a microscope. We noted the size of the cells, and vascularization, and any specialized cells. The next step was to examine different prepared slides of fungi and identify which of the three fungi groups they belonged to. The third step in lab was to create a Berlese Funnel to collect invertebrates. To do this, we took a small plastic tube and filled it with 25mL of ethanol. We taped a funnel to this tube and laid a piece of black mesh over the funnel’s hole. We then stuffed the funnel full of our leaf litter samples and hooked the entire contraption to a pole underneath a light source. The Berlese Funnel will catch invertebrates in the ethanol below, while the black mesh will keep the leaf litter from contaminating the ethanol. The entire class’s Berlese Funnels were then covered in aluminum foil to keep the light locked in.
Data and Observations
Since it was wintertime, there were few plants alive in our transect. We took a leaf from a medium sized tree in the bottom right corner of the transect whose leaves grew in patterns of three from a central point. We took another sample from a small tree in the top right that had only dead leaves. This tree’s leaves seemed to only grow one at a time, each from a different starting point. We gathered samples from two different kinds of monkey grass that grew near the rock path in the middle of the transect. One type had very long, thin, light green leaves, and the other had shorter, thicker, darker green leaves. Both grasses had leaves that originated from one center in the ground. Our last sample was from a purple plant that had layers of leaves growing up a central stalk. This plant came from the bottom left corner of the transect. Though it was difficult to tell in the winter when no evidence of flowering exists, we belief most of our plants were angiosperms, or flowering plants. The purple plant still had evidence of a flowering body on its head and grasses are known to be angiosperm. The two trees were hardeItalic textr to tell, but since the smaller tree was clearly seasonal, it is safe to assume it is an angiosperm. The live tree, however, was harder to tell. Since it had visible veins and broad leaves, we again assumed it was an angiosperm. Below is a map with the locations of the plants circled.
Below are pictures of the five different plants in the transect and a close up picture of the sample we took from each plant. All microscopic pictures were taken at 10x.
1: Monkey Grass 1
2: Green Tree
3: Monkey Grass 2
4: Purple Plant
5: Dead Tree
When we examined the cross sections under the microscope, we noticed that the monkey grass had the clearest vascularization. The light green monkey grass had wide, vertical veins evident on the cellular level, and the darker green grass had smaller veins. The purple plant was multicolored on a cellular level, with each cell a different shade of purple, red, orange, or green. Below is a table of our findings.
We then examined fungi. I looked at agaricus bisporus, rhizopus stoloniger zygospore, and rhizopus nigricons sporangia. Agaricus bisporus, or the common mushroom, is a basidiomycota gunfi because of its special gill-like structures on its underside that allow for sexual reproduction. Rhizopus stolonifer zygospore, or black bread mold, is a zygomycota because it reproduces by creating a zygospore. These fungi structures called sporangia that contain spores. These spores are the way that zygomycota fungi reproduce and are also the reason that black bread mold has its distinct black color. Finally, rhizopus nigricans sporangia is also a zygomycota. Below is a drawing and a picture of rhizopus nigricans sporangia. The bulb-like structure shown is the sporangia.
Conclusions and Further Study The diversity of plants is astounding. After analyzing only five plants from the Wildlife Habitat transect, the differences in size, structure, and vascularization were plain. Each plant provides different resources to its ecosystem and requires different conditions to survive. This lab provided a greater understanding of the diversity of the world of plants and their impacts on an ecosystem.
2/2 EB
Lab 3: Bacteria in Transect 2
Introduction
The core goal of this lab was to observe the bacteria that grew in transect 2 and to understand antibiotic resistance in bacteria. After plating serial dilutions of samples from our Hay Infusion onto both nutrient and antibiotic-laced plates, my group hypothesized that the plates with antibiotics on them will have less bacteria growing on them. This is intuitive because bacteria will have a harder time growing in environments with particles designed to destroy them. We also predict that there will be no Archaea species in our plates. Archaea typically live in very harsh environments, and the wildlife habitat of transect two is not harsh at all. Therefore, though this lab described different Archaea species, we do not anticipate finding any of them in our samples.
Materials and Methods
First we observed our Hay Infusions to see how they had changed over the course of the week. After taking several pictures of the infusion, we moved on to analyzing the bacteria plates. We collected our plates and observed the color and size of both the nutrient and the tetracycline (henceforth referred to as tet) plates. We counted, as best we could, the number of colonies on each plate and recorded the information. We next created two wet slides of the bacteria. We took one sample from the clear bacteria growing in the 10-9 nutrient plate and one sample from the orange bacteria growing in the 10-3 tet plate. Using a metal loop, we put the samples on a slide and observed them as best we could. Without staining, however, it was difficult to find and see the bacteria clearly. The next step, then, was to stain the bacteria. We did this by taking four chosen slides (10-3 and 10-9 in both nutrient and tet plates) and sealing the bacteria into place by passing it several times over a flame. After the bacteria was heat fixed, we put the slides over a dying bin and washing them with crystal violet die, iodine mordant, alcohol, and safranin stain. Between each stain, we washed the entire slide with water to clear the previous stain away. Once the bacteria was stained, washed, and dried, we used microscopes to exam each sample and determine whether it was gram positive or negative. If the color remained in the bacteria it was gram positive, and if there was no color, it was gram negative. Once the observation was complete, we took four samples from the agar plates again to prepare for PCR. We transferred the bacteria into 100 microliters of water, incubated it at 100°C for 10 minutes, and centrifuged it for 5 minutes. We then added water to a PCR tube to dissolve the bead and poured the supernatant from the centrifuged tubes into the PCR tube. We then placed it in the PCR machine.
Data and Observations
The Hay Infusion appeared to have settled into two distinct groups. The heavier materials had fallen to the bottom and formed an ecosystem of their own, while the water throughout the rest of the jar appeared much less dense. The Hay Infusion had increased drastically in smelliness, stinking of rotting animals. Below are pictures of the Hay Infusion.
Below is a picture of the data we recorded from our agar plates. The nutrient agar plates appeared to have two different kinds of bacteria. The first was bright orange, shiny, and abundant in both the tet and the nutrient plates. The second was clear, smaller, and much harder to see. Though the tet and nutrient plates had similar numbers of the orange bacteria in each, the clear bacteria only showed up in the nutrient plate. This suggests that the clear bacteria was not resistant to tetracyclinee, but that the orange bacteria was almost completely resistant, since it showed no decreases in number between the two plates. This provides support for our hypothesis that the nutrient plates would have more bacteria in them.
Tetracycline is an antibiotic that works as a protein inhibitor. It blocks tRNA from attaching to the A site in protein synthesis (Chopra and Roberts 2001). It is particularly potent to bacterial cells because they pump tetracycline into their cytoplasms, while many eukaryotic cells do not. Tetracycline more easily navigates the cell walls of gram-negative bacteria. Though originally the antibiotic was affective on many bacteria, gram positive or negative, exposure to the antibiotic has allowed many organisms, especially the gram positive, to become resistant (Chopra and Roberts 2001).
Table 1
10-3 Nutrient Plate
10-5 Nutrient Plate
10-7 Nutrient Plate
10-9 Nutrient Plate
10-3 Tet Plate
10-5 Tet Plate
10-7 Tet Plate
10-9 Tet Plate
The wet mount samples were difficult to find since they weren’t stained. The sample of the clear bacteria was much larger and easier to see than the sample of the orange bacteria, shown as the fuzzy white dot below. None of the cells seemed motile.
Wet Mount of 10-3 Tet Orange Bacteria
Wet Mount of 10-9 Nutrient Clear Bacteria
Below are pictures and a chart of the results of our gram staining.
Table 2
10-3 Nutrient Orange Bacteria (40x)
10-9 Nutrient Clear Bacteria (40x) - Note the multiple different shapes the bacteria takes on.
10-3 Tet Orange Bacteria (40x)
10-9 Tet Bacteria (40x) - There are two pictures of the -9 tet bacteria to show the differences in appearance.
Conclusions and Further Study
This lab revealed that at least two different kinds of bacteria were living in our transect. One with small and clear colonies and the other with large orange colonies, both bacteria were gram positive. However, only the clear bacteria was affected by the antibiotic. This data supports our hypothesis that fewer bacteria would thrive in the tet plates and our hypothesis that no Archaea species would live in our transect. There was a core source of error in our data, however. The staining process was messy, making it difficult to tell whether cells truly were gram positive or gram negative. Future studies should address this issue by increasing the washing and cleaning time during the staining process.
Works Cited
Chopra, I., & Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiology and molecular biology reviews, 65(2), 232-260.
1/25 EB
Lab 2: Discovering Protist Habitats
Introduction
Despite common opinion, ecosystems can be as large or as small as the environment allows. Though a 20 by 20 meter transect is an ecosystem in its own right, there are much smaller, more specified ecosystems and niches throughout the entire area. Even the niche from the surface of our Hay Infusion differs from the niche on the bottom. The purpose of this week’s lab was both to understand these nuances of ecosystem dynamics and to determine which protists resided in our whole transect’s ecosystem. Without a firm understanding of the microscopic ecosystem of our area, we cannot accurately study the transect as a whole.
Materials and Methods
The core goal of this lab was to identify different protists that lived in the Hay Infusion made from a sample of our transect’s dirt. We first practiced identifying protists using a dichotomous key. Once we felt comfortable finding and identifying protists, we moved on to studying protists from our Hay infusion. We took samples using a transfer pipette from both the surface and the bottom of the Hay Infusion, and placed them in separate flat disk containers. We took a sample from approximately 1cm from the bottom in the middle of the jar, and one sample right below the growths on the surface. We tried to take samples from the growths themselves, but they were too large to be transferred by the pipette. With two lids of water representing the two different niches, we then made three wet mounts of each niche. Each member of our group took one wet mount from the top and one from the bottom to observe. We tried to find three specimens from the top and three from the bottom using the dichotomous key to identify any cells we found. For each specimen, we recorded its appearance, size, and scientific name. At the end of the lab, we created a serial dilution to prepare for our bacteria lab the next week. We had four plates of agar and four plates of agar plus an antibiotic, tetracycline. We then took 100 microliters of our Hay Infusion (thoroughly mixed), and put it into a tube with 10mL of broth. From there, we took 100µL and added it to a different tube with broth. From the second tube we took 100µL and added it to a third, and from the third we took 100µL and added it to the fourth. This is a serial dilution. Each successive tube is more diluted than the one before it. Below is a picture detailing the methods for this dilution.
Data and Observations
The Hay Infusion smelled like musty dirt and rotting sewage. A film had grown on the surface, with most of the dirt and plant matter settling to the bottom. A picture of the jar is below.

Our group found six different protists in the Hay Infusion. However, possibly because we were unable to pipette water from the surface due to the film growth and instead had to draw from directly under the film, there was far more diversity in the bottom niche than on the top. We found four different protists from the bottom, but were only able to identify two different protists from the top, both paramecia. Below is a table detailing our findings and drawings of each protist.
Perhaps the reason so few organisms lived in the surface niche was because of the film of life that grew on the top of the water. Sunlight and other resources might have been monopolized by the growth on top, leaving less for other organisms that might usually thrive in a surface level niche. Organisms on the bottom might prefer different conditions and require different resources than those on top, explaining why such diversity appeared there. For example, there was more plant matter on the surface of the culture than on the bottom. Therefore, heterotrophic cells, like paramecia, would be more likely to thrive on the surface, where food is readily available, than on the bottom. This is shown by our results. We found only heterotrophic species on the surface, whereas 3 out of the four bottom protists were autotrophs.
Though many protists are single-celled and relatively simple, they fit all the requirements for life. Take Gonium, for instance. Though it simple and non-motile, it fits all five of Freeman’s needs for life: made of cells, uses energy, processes information, replicates, and is a product of evolution (Freeman 2008). Gonium colonies typically contain 16 or less cells. They are cells because they are surrounded by a phospholipid bilayer. They use energy through photosynthesis, which is easy to ascertain by observing their green color. They can reproduce through either sexual or asexual reproduction, which shows that they are a product of evolution. Farther back in their evolutionary line, cells only reproduced through asexual reproduction (Clamydomonas is an example). Gonium, however, is an intermediary step between asexual and sexual reproduction, capable of both. Finally, they can process information, shown by the fact that they have and use DNA (Bentley 2015).
Conclusion and Further Direction
Our Hay Infusion fostered a plethora of different protist species in the one week it had to grow. From photosynthesizing, single celled organisms to large colonies of cells, to motile heterotrophic cells, protists of all natures flourished in the culture. If left to grow even longer, the culture certainly would have gained even more life. However, the diversity of this life is questionable. In the beginning stages of growth, many different species could thrive in the culture, since space would appear unlimited. As the cells continued to grow and reproduce, however, competition for resources would increase. Perhaps, if left for several months, only the best-suited protists would remain in the culture, having defeated other protists in the fight for resources. On the other hand, a longer time lapse would allow more cells to enter the culture through the air. As different students entered and exited the classroom, different particles could be deposited into the culture. This would lead to greater diversity. Without experimenting, it is impossible to know how the Hay Infusion would change through time. Future labs should test the hypothesis that extended time would lead to either an increase or decrease in diversity in the culture.
References
Bentley, Meg. 2015. A Laboratory Manual to Accompany: General Biology II. Department of Biology, American University.
Freeman, Scott. 2008. Biological Science. Pearson Benjamin Cummings: San Francisco. 130. Print.
1/22/14 EB
Lab 1: Observing a Transect
Intro
Observation, simple as it may sound, is key to understanding science. No theories, hypotheses, or even scientific questions can emerge without this first vital step of the scientific method. This lab’s core goal was to collect observations about a specific transect on AU’s campus, namely the Wildlife Habitat transect, located between Cassel Hall and Hughes Hall. My group observed, drew, sampled, and analyzed the 20 by 20 meter plot in order to grasp how organisms within an ecosystem interact and develop. The data collected from this initial observation will be used throughout this semester’s study of campus ecosystems.
Materials and Methods
For this lab, the most important material was simply sensory information. Aside from the tube used to collect a dirt sample, all data was collected from observing, listening, and smelling a given transect. We first drew a map of the plot, including both the area within the transect and a few meters outside our area to show the boundaries. We then scrutinized each part of the transect, listing each biotic and abiotic element we found. Finally, we collected a sample of dirt representative of that area in a tube to use for Hay Infusions. The second part of lab was preparing the Hay Infusions for next class. We added .1 grams of powdered milk and 10 grams of dirt to a large jar filled with 50mLs of water in order to create an ecosystem where protists could thrive. After shaking the mixture to ensure the milk and dirt were properly mixed, we left the jars, lids off, for a week to allow the ecosystem time to grow.
Data Collected
Below is both a drawing and digital picture of my group’s transect, the wildlife habitat ecosystem, or transect 2.

Our transect was located on the North side of campus, on a slight slope. Aside from the slight downward tilt, the transect was mostly flat, with few bumps or small hills. The soil appeared hard and dry, but was surprising moist and loose once we began collecting samples. We found that our transect contained diverse biotic elements, but relatively uniform abiotic elements. The abiotic factors we documented were a piece of crumpled tissue paper, a few large boulders on the periphery of the transect, a row of smaller rocks dividing the transect, and lots of dirt and mulch throughout. Most of the transect, in fact, was covered in brittle dirt or scattered with wood chip mulch. When we came to biotic factors, we found more diversity. For plants, there were four trees, two of them big, one medium, and one small, that created the boundaries for our transect. There were several clumps of shrubs, some in the south with numerous purple leaves and some towards the north along the rocks that were stick-like and bare of leaves. Small clumps of monkey grass grew right next to the rocks as well, and fallen leaves scattered the ground. Since we were out late at night, there were few bird species, but observing the transect during the day revealed numerous sparrows. Though not seen, our group heard and felt at least one rat scurrying in the transect. We saw no bugs in the transect, but their absence could be due to the cold winter weather.
Conclusions and Future Directions
Since this lab did not attempt to test any hypothesis, there are no conclusions regarding any outcomes of our observations. However, the data we collected showed that the our transect contained a diverse assortment of plants, a small number of animals, and a few different abiotic components. The area was relatively litter-free, containing only one piece of trash. This cleanliness combined with the surprisingly moist soil allowed the transect to live up to its name of ‘wildlife habitat.’ The soil sample we collected should be an accurate representation of the transect as a whole, though the cold of winter, which left most of the plants in the transect deadened, might have affected the overall protist population of the region. Future tests should gather samples from numerous different seasons to determine the affect of cold weather on protist life.