User:Danyan Tang/Notebook/Biology 210 at AU: Difference between revisions
Danyan Tang (talk | contribs) No edit summary |
Danyan Tang (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
'''PCR for 16S sequencing March 4th 2015''' | |||
Purpose: This lab was to use the PCR products to identify our bacteria species in agar plant from the hay infusion culture. | |||
Material and Methods: | |||
1. Transferred a single colony bacteria form the agar plates was to 100 μL of water in a sterile tube. | |||
2. To incubate at 100 °C for 10 minutes in a heat block. | |||
3. Centrifuged the samples for 5 minutes at 13,400 rpm. | |||
4. During the centrifuge, 20 μL of primer and water mixture was added to a PCR tube and mixed until the PCR bead was dissolved. | |||
5. 5 μL of the supernatant form the centrifuged samples was transferred to the 16s PCR reaction and placed in the PCR machine. | |||
6. After one week, the PCR products were ran out on an agarose gel and purified the DNA for sequencing. Then selected two best sample. | |||
7. After we got the results from GeneWiz and Genomic Blas, we used the data to identify the bacteria species. | |||
Results: | |||
The samples we selected in the agarose gel, they were 10^-5 dilution without tetracycline and the 10^-3 dilution with tetracycline. | |||
[[Image:Adjfnajcnee.jpg]] | |||
Raw Sequences: | |||
MB43 GGNNNNNNNNNNNNNNNNANNNTGCAGTCGTACAGGTAGCCGTAANTTGCTCTCGGGTGACGAGTGGCGGACGGGTGANT AATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAATACCGCATAACGTCGCAAGACCAAAG AGGGGGACCTTCGGGCCTCTTGCCATCAGATGTGCCCAGATGGGATTAGCTAGTAGGTGGGGTAATGGCTCACCTAGGCG ACGATCCCTAGCTGGTCTGAGAGGATGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGT GGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTATGAAGAAGGCCTTCGGGTTGTAAAGTACTT TCAGCGGGGAGGAAGGTGTTGTGGTTAATAACCGCAGCAATTGACGTTACCCGCANAANAAGCACCGGCTAACTCCGTGC CAGCANCCGCGGTAATACGGANGGTGCAAGCGTTAATCGGNAATTACTGGGCGTAAAAGCGCACGCAGGCGGTCTGTCAA GTCGGATGTGAAANTCCCCCGGGCTCAACCTGGGAACTG | |||
MB44 NNNNNNNNNNNNNNNNCNNNNNNNNGACAGCCGAGCGGTAGAGATCTTTCGGGATCTTGAGAGCGNGCGNTACGGGTGCG GANCNNNTGTGCAACCTGCCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATATTGAATGGCA TCATTTGATATTGAAAACTCCGGTGGATAGAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAG TCAGTGATCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGC AGTGAGGAATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGACGACGGCCCTATGGGTTGTAAACT TCTTTTGTATAGGGATAAACCTTTCCACGTGTGGAAAGCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCA GCAGCCGCGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGATCTGTAAGTCA GTGGTGAAATCTCATAGCTTAACTATGAAACTGCCATTGATACTGCAGGTCTTGAGTAAANGTANAAGTGGCTGGAATAA GTANTGTANCGGTGAAATGCATAGATATTACTTANAACACCNATTGCGANNCAGGTCACTATGNTTTAACTGACGCTGAT GGACGAAAGCGTGGGGAGCGAACNGGATTANATACCCTGGGTAGTCCACGCCGTAAACNATGCTAACTCGTTTTTGGNCT TTAGGGTTCAGANACTAAACNAAAGTGATNAGTTAAGCCNCCTGGGGANTACGTTCGCAAGAATGAAACTCANAGGAATT GAACGGGGGCCCGCACACCGGGGGATTATGTGGTTTANTNNNATNANTCNCANGGAACCNTACCANGCTAAATGGGNATT GANGGGTNNNNANTAGACTTTCTTCNANNNTTTCAANGNNCTNCATGGGTGGNNGNGNGCTNNNGCNNNNAAGNNNNNNN N | |||
species ID: | |||
MB43- Enterobacteriaceae bacterium S2A (96% match). | |||
MB44- Uncultured bacterium clone (89% match). | |||
Conclusion: | |||
The MB43, which is Enterobacteriaceae Bacterium S2A, corresponds to the result we obtained on the agar plants, which was gram negative. From the reference, this bacterial species is gram negative which means match our date from gram staining lab. The 16s sequencing of the bacterial sample of MB44 has the lower matching percentage and poor quality which means it doesn’t have enough information for further studying and we can not compare this to our results from previous gram staining procedure. | |||
---- | |||
'''Lab 6 Embryology&Zebrafish Development Feb 25 2015 ''' | '''Lab 6 Embryology&Zebrafish Development Feb 25 2015 ''' | ||
Revision as of 23:36, 4 March 2015
PCR for 16S sequencing March 4th 2015 Purpose: This lab was to use the PCR products to identify our bacteria species in agar plant from the hay infusion culture.
Material and Methods:
1. Transferred a single colony bacteria form the agar plates was to 100 μL of water in a sterile tube. 2. To incubate at 100 °C for 10 minutes in a heat block. 3. Centrifuged the samples for 5 minutes at 13,400 rpm. 4. During the centrifuge, 20 μL of primer and water mixture was added to a PCR tube and mixed until the PCR bead was dissolved. 5. 5 μL of the supernatant form the centrifuged samples was transferred to the 16s PCR reaction and placed in the PCR machine. 6. After one week, the PCR products were ran out on an agarose gel and purified the DNA for sequencing. Then selected two best sample. 7. After we got the results from GeneWiz and Genomic Blas, we used the data to identify the bacteria species.
Results:
The samples we selected in the agarose gel, they were 10^-5 dilution without tetracycline and the 10^-3 dilution with tetracycline.

Raw Sequences: MB43 GGNNNNNNNNNNNNNNNNANNNTGCAGTCGTACAGGTAGCCGTAANTTGCTCTCGGGTGACGAGTGGCGGACGGGTGANT AATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAATACCGCATAACGTCGCAAGACCAAAG AGGGGGACCTTCGGGCCTCTTGCCATCAGATGTGCCCAGATGGGATTAGCTAGTAGGTGGGGTAATGGCTCACCTAGGCG ACGATCCCTAGCTGGTCTGAGAGGATGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGT GGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTATGAAGAAGGCCTTCGGGTTGTAAAGTACTT TCAGCGGGGAGGAAGGTGTTGTGGTTAATAACCGCAGCAATTGACGTTACCCGCANAANAAGCACCGGCTAACTCCGTGC CAGCANCCGCGGTAATACGGANGGTGCAAGCGTTAATCGGNAATTACTGGGCGTAAAAGCGCACGCAGGCGGTCTGTCAA GTCGGATGTGAAANTCCCCCGGGCTCAACCTGGGAACTG
MB44 NNNNNNNNNNNNNNNNCNNNNNNNNGACAGCCGAGCGGTAGAGATCTTTCGGGATCTTGAGAGCGNGCGNTACGGGTGCG GANCNNNTGTGCAACCTGCCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATATTGAATGGCA TCATTTGATATTGAAAACTCCGGTGGATAGAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAG TCAGTGATCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGC AGTGAGGAATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGACGACGGCCCTATGGGTTGTAAACT TCTTTTGTATAGGGATAAACCTTTCCACGTGTGGAAAGCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCA GCAGCCGCGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGATCTGTAAGTCA GTGGTGAAATCTCATAGCTTAACTATGAAACTGCCATTGATACTGCAGGTCTTGAGTAAANGTANAAGTGGCTGGAATAA GTANTGTANCGGTGAAATGCATAGATATTACTTANAACACCNATTGCGANNCAGGTCACTATGNTTTAACTGACGCTGAT GGACGAAAGCGTGGGGAGCGAACNGGATTANATACCCTGGGTAGTCCACGCCGTAAACNATGCTAACTCGTTTTTGGNCT TTAGGGTTCAGANACTAAACNAAAGTGATNAGTTAAGCCNCCTGGGGANTACGTTCGCAAGAATGAAACTCANAGGAATT GAACGGGGGCCCGCACACCGGGGGATTATGTGGTTTANTNNNATNANTCNCANGGAACCNTACCANGCTAAATGGGNATT GANGGGTNNNNANTAGACTTTCTTCNANNNTTTCAANGNNCTNCATGGGTGGNNGNGNGCTNNNGCNNNNAAGNNNNNNN N
species ID:
MB43- Enterobacteriaceae bacterium S2A (96% match). MB44- Uncultured bacterium clone (89% match).
Conclusion: The MB43, which is Enterobacteriaceae Bacterium S2A, corresponds to the result we obtained on the agar plants, which was gram negative. From the reference, this bacterial species is gram negative which means match our date from gram staining lab. The 16s sequencing of the bacterial sample of MB44 has the lower matching percentage and poor quality which means it doesn’t have enough information for further studying and we can not compare this to our results from previous gram staining procedure.
Lab 6 Embryology&Zebrafish Development Feb 25 2015 Purpose: We set up an experiment with zebrafish by using different concentration of ethanol to learn the stage of embryonic development and see how environmental conditions affect embryonic development.The hypothesis is if ethanol is added to a zebrafish embryo, the development of the zebrafish will be affected. We predict that the ethanol will cause smaller eye diameter, slower heart rate and a higher death rate in zebrafish.
Material and methods: We set up three different concentration of ethanol in three separate petri disses. One was the control group contained 20mls deerpark water, the other were a dilution of 0.75% of ethanol and 0.325% of ethanol. Then, we selected 20 healthy translucent embryos and transferred to each solution by using transfer pipette. In the next 14 days, we will observe these three dishes under a microscope and record their change, also remove some amount of solution.
Data and Observation:Observation of DAY 0-DAY 4 Table 1: Observation of water
| ' | ' | Day 0 | ' | Day1 | ' | Day4 | ' | ' | ' | Day7 | ' | ' | Day11 | ' | ' | Day14 | ' | ' |
| Dead eggs | 0 | 0 | 10 | 2 | 6 eggs | |||||||||||||
| Number surviving | 20 | 20 | 10 | 6 eggs,2 hatched | 2 hatched | 2 hatchlings | ||||||||||||
| remove solution | N/A | N/A | 10ml water | 10ml water, dead eggs | 10ml water,dead eggs | N/A | ||||||||||||
| add solution | 20ml water | N/A | 25 | 10ml fresh water | 10ml fresh water | N/A | ||||||||||||
| length | N/A | N/A | N/A | 4mm | 4mm | One is 4mm, the other 4.5mm with fins | ||||||||||||
| developmental stage | dome to zygote | Oblong to Sphere | 2 fish~36hour, others are bud stage | 20 somite, fish are 48 hour hatched | hatched | hatched | ||||||||||||
| movement | N/A | N/A | fish are swiftly moving through solution using tales | fish are swiftly moving using tails | subtly movement, eyes move a lot | tail,slow eys move a lot | ||||||||||||
| eye diameter | N/A | N/A | 0.02 mm | 0.05 mm, head is 0.75 mm | 0.05 mm, head is 0.75 mm | 0.05mm | ||||||||||||
| heart rate(beats/min) | N/A | N/A | N/A | 66 | 70 | 78 |
Table 2: Observation of 0.75% Ethanol
| ' | ' | Day 0 | ' | ' | Day 1 | ' | Day 4 | ' | ' | ' | Day7 | Day 11 | ' | ' | Day 14 | ' | ' |
| Dead eggs | 0 | 0 | 9 | 8 | 0 | 3 hatchlings | |||||||||||
| Number surviving | 20 | 20 | 11 | 3 hatched | 3 hatched | 0 | |||||||||||
| remove solution(ml) | N/A | N/A | 10 | 10ml | 10ml | N/A | |||||||||||
| add solution(ml) | 10ml 1.5% enthanol +10ml water | N/A | 25 | 10ml | 5ml ethanol+5ml water | N/A | |||||||||||
| length | N/A | N/A | N/A | 4mm | 4mm | 4mm | |||||||||||
| developmental stage | Dome to zygote | oblong to sphere | 2 fish~36hour, others are bud stage | 48hours | hatched, greener than the control group | black | |||||||||||
| movement | N/A | N/A | fish are swiftly moving through solution using tales | using tails | quick and random | N/A | |||||||||||
| eye diameter | N/A | N/A | 0.02mm | 0.025mm | 0.025mm head is 1mm | N/A | |||||||||||
| heart rate | N/A | N/A | N/A | 36 | 45 | ||||||||||||
Table 3: Observation of 0.325% Ethanol
| ' | ' | Day 0 | ' | ' | Day 1 | ' | Day 4 | ' | ' | ' | Day 7 | ' | ' | ' | Day 11 | ' | ' | ' | Day 14 |
| Dead eggs | 0 | 0 | 3 | 7 | 3 | 2 eggs dead, 1 hatching | |||||||||||||
| Number surviving | 20 | 20 | 17 | 4 fish.6 eggs | 4 hatched,2 eggs | 3 hatchings | |||||||||||||
| remove solution(ml) | N/A | N/A | 10 | 10ml | 10ml dead eggs | N/A | |||||||||||||
| add solution(ml) | 5ml 1.5% enthanol +15ml water | N/A | 25 | 10ml | 2.5ml ethanol, 7.5ml water | N/A | |||||||||||||
| length | N/A | N/A | 0.8mm | 5mm | 5mm | 5mm | |||||||||||||
| developmental stage | Dome to zygote | oblong to sphere | 2 fish~36hour, others are bud stage | 48 hours hatched | hatched,darker body cavity | hatchling with larger body cavity | |||||||||||||
| movement | N/A | N/A | fish are swiftly moving through solution using tales | tails | quicker than control group, random movement | rapid random movement, cant stay still | |||||||||||||
| eye diameter | N/A | N/A | 0.02mm | 0.25mm | 0.05mm head is 1mm | 0.05 eye diameter, head is 1.5 mm | |||||||||||||
| heart rate | N/A | N/A | N/A | 36 | 36 | 60 |
Conclusion: At this moment, our hypothesis is invalid, we still have to wait more days because most of embryo still need time to hatch out. At this time, we can see that the result shows the highest survival rate in the 0.325% solution and we haven't got any heat rate,therefore we can not make conclusion now.
A.T
2.20.15 Very good notebook entry. Nice description of experimental procedures and data. I think you should have drawn your own food web. SK
Lab 5 Invertebrates Feb 18 2015 Purpose: This lab was designed to understand the importance of how invertebrates function and how simple systems evolved into complex systems.by investigating the invertebrates collected from the Burlese Funnel from the transect.
Material and methods: 1. To observe the provided slides which included Planaria, nematodes and Annelida by using a dissecting scope. To examine the type of movement for each organism. 2.Observed the burls funnel by using dissecting scope, separated the liquid from the funnel into two petri dishes. 3. To identify five vertebrates and determined their classification. also considered the biotic and abiotic characteristics of the transect that would benefit each species and constructed a food web.
Data and observation: When we observed the provided slides under a dissecting scope Planaria was flatworm solid bodies which moved smoothly and gliding motion. The nematodes were had round bodies and their movement were random which their major muscles are down dorsal and ventral sides. Finally, Annelida were a fully lined fluid-filled which called coelomate, moved with a rippling effect which facilitates movement by peristalsis.Then, we observed the funnel and found three organisms.(#1,2,3) As shown in the table, The largest organism was the worm which measured 2mm, the second was the beetle with a measurement of 1.5mm. The smallest size was the spider, measured by 0.5mm.
Vertebrates in Transect 2: Wildlife Habitat. by Claudia J.Woloshchuk
by Claudia J.Woloshchuk
Food web 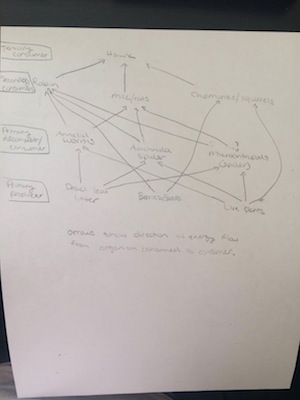 by Claudia J.Woloshchuk
by Claudia J.Woloshchuk
Conclusion: After we observed invertebrates that found in the Berlese funnel, we could understand how important are they, how they function in our transect and the biodiversity of organism. From the food web, we could understand the relationship between the organisms and each organism has their own position which benefits each other.
AT.
2.20.15 Good description of methods. Need to include some more text in results and conclusion sections describing your plants and the pictures. SK
Lab 4.Plantae and Fungi Feb 11th 2015
Purpose:To understand the characteristics and diversity of plants by analyzing their vascularization, specialized their structure and mechanisms of reproduction.Also,to appreciate the function and importance of fungi by observing three different types of organism under microscope.
Material and methods: 1.We collected five different plants in a ziploc bag from our transect area and observed them under microscope,then made a table about the information of them. 2.Observed the slides of Bryophyte moss, Mnium, and an angiosperm, Lilium to understand the diversity of plants. 3.In order to understand the mechanisms of reproduction, we dissected the flower of lily and observed all the parts of it 4.In order to appreciate fungi, we observed three different types of fungi. 5.We set up the berlese funnel to collect invertebrates to prepare the next lab. 6. Poured 25ml of the 50:50 ethanol/water solution into the 50ml conical tube and fitted a piece of the screening material into the bottom of the funnel. 7.Put the leaf and soil on the top of funnel and set up the funnel on the ring stand and placed the 40 watt lamp with the incandescent bulb about 1-2 inches from the top of the funnel. Leave the funnel for one week. Data and Observation: Five different types of plants from transect area were observed and analyzed by using a table(#1,2,3,4,5,6) and three types of fungi were observed which were Sporangia Rhizopus nigrican, a mushroom, and Rizopus stolonifer.(#7,8.9)
photos by Corina C. Velazco
Conclusion:After this lab, we can understand our transect area more deeply and from our collect samples, we can understand they are monocots or dicots, structure and mechanisms of reproduction.
2.9.15 Very good notebook entry. The images of the results tables are small which makes it difficult to read the text. SK
Lab 3.Microbiology and identify bacterial with DNA sequences Feb 4th 2015 Purpose: This lab is designed to understand how bacteria behaves in ecosystem and the characteristics of them,to observe antibiotic resistance and to differentiate microorganisms by using the techniques of staining. In addition,to identify bacteria by using DNA sequence which we will continue on Lab 4.
Material and methods: 1.To observe the Hay Infusion Culture and recorded any change of appearance and smell. 2.Observed the eight agar plates we made last week and recorded the colonies we counted. 3.To make four wet amount of agar plate which were two samples with Tet and 2 samples without Tet namely and examined under microscope. To take the sample, we sterilized the loop first and scraped up a tiny amount of colony then placed it after added a drop of water on the slide. proceeded to the 100x and added a drop of oil on it. 4. After the procedure of observe sample under the microscope, we did the gram staining to see whether they were positive or negative which means contains more or less peptidoglycan. Recorded them on the lab book. 5. To set up a PCR for 16s sequencing. Transferred one single colony to a sterile tube that contained 100µl of water and then incubated in 100°C for 10 minutes and then centrifuged for 5 minutes at 13,400rpm. After centrifugation, 5µl of the supernatant was placed in the PCR tube that had the PCR bead and then added the 20µl of the primer.Repeated this step for 3 more samples, one with Tet and two without Tet.
Data and Observation: There will not any Archaea species have grown on the agar plate, because Archaea species tend to be extremophile and the environment in the plate can not offer the conditions to grow .The appearance of our culture this week was different than last week. most of the water had evaporated but the smell was not as bad as last week andthe berries resemble shriveled grape. It is hypothesized that the smell has changed because the water became less therefore the organisms have died. For the eight agar plates, the plates without Tet had more colonies that the plates with Tet and the result of 100-fold serial dilution shows below.(plates without Tet#1&2 plates with Tet#3&4 and photos by Claudia J. Woloshchuk table of lab book#5) From the observed plates, we could see the plates that with Tet had less colonies means less bacterias and the organisms we found from our culture do not have antibiotic resistance. The mechanisms of action for Tet is" Entry of these agents into susceptible organisms is mediated both by passive diffusion and by an energy-dependent transport protein mechanism unique to the bacterial inner cytoplasmic membrane. Nonresistant strains concentrate the tetracyclines intracellularly. The drug binds reversibly to the 30S subunit of the bacterial ribosome, thereby blocking access of the amino acyl-tRNA to the mRNA-ribosome complex at the acceptor site. By this mechanism, bacterial protein synthesis is inhibited."(Guzman 2008) The way to differentiate microorganisms is on staining characteristics.(#6)
conclusion: Obviously, after what we did, it prove that the organism with antibiotic would grow less than the one without antibiotic, i think there is why antibiotic has been used for serious illness.However, heavy use of antibiotic will contributes to the problem of antimircobial resistance so there still have argument of using this.
AT
2.4.15 Good notebook entry. Identified protists found in Hay Infusion and location in Hay Infusion. Included good pictures and organized well. SK
Lab 2. identifying algae and protists Jan 28 2015 Purpose: To identify different unknown organisms by using a dichotomous key and use this to understand the characteristics of protists and algae. Also to identify protists and algae in the Hay infusion Culture was made from our assigned transect.
Material and methods: 1.Observed a wet mount of known organisms under microscope and used the dichotomous key to identify them. 2.To observe our culture by taking from 2 different niches,we took them from top and bottom separately.Then made a wet mount for each of the samples and identified protist and algae from the Hay Infusion Culture by using the dichotomous key. 3.prepared a total of eight petri dishes by using sterile broth,mixture of Hay infusion culture,nutrient agar and nutrient agar plus tetracycline.Placed all of them at room temperature and will use them for lab 3.
Data and observation: when we observed the Hay infusion culture we made last week, we could smell the water likes stinking sewers and it started to evaporate. We could see the moss and soil were in the bottom and there were a couple of berries and wood on the top of muddy and turbid water. There is no apparent life like molds or green shoots on the top the liquid.(#1&2) We took samples from the top and bottom, colpidium and gonium were found from the top and also found colpidium from bottom. Gonium was about 20µm, appears in green color with disc-shaped cells.(#3&5) Colpidium 25µm, appears colorless and oval-shaped. It exhibited motility because of cilia.(#4&5) Prepare for lab 3. (#6)
1.

 photo by Corina.C
4.
photo by Corina.C
4. Photo by Corina.C
5.
Photo by Corina.C
5.

Conclusion and future direction: After lab 2, we understand how to identify different organisms using the dichotomous key. However, only one week is not enough time let the our jar to grow, if we put the culture for a long time period they might reproduce then we can find our more organisms. In the future, we can supply our culture some nutrient in order to keep them for future investigation.
A.T
1.27.15 Very good first entry. Try to use past tense and describe what you did rather than what was in the lab manual instructions. Try to rotate the photos if possible. SK
Lab 1. Biological life at AU Jan 15 2015
Purpose: 1.Observe 3 types of green algae using microscope and depression slides and understand how natural selection leads the evolution. 2. To observe and analyze a assigned transect at the AU campus.We will use the biotic and abiotic as samples to examine through the serval weeks for this semester and discuss the relationships between species in ecosystem.
Material and methods:1.(for the first part of lab) observe 3 types of green algae under the microscope which were chlamydomonas, gonium and volvox. Analyze them and fill out the information on the notebook. 2.(for the second part of lab) The TA gave each one a number to form a group of three assigned to each transect. our group went to the transect to take the sample with a 50mL conical tube.Back to the lab, we weighed out 11.22g of soil sample transferred them to a jar and added 500ml Deerpark water and 0.14g dried milk powder to make Hay infusion culture. Mix the jar for 10 second and placed them without lid for next week.
Data and observation:1.examine 3 green algae group,chlorophyta--the volvocine line. (#1)2. The 20 by 20 meter assigned transect was located on the North side of campus on the side where Hughes Hall and McDowell Hall are adjacent to each other.(#2) There are some abiotic and biotic components. abiotic:rocks, sunlight, shade from building,bench, soil and snow. biotic: tree, moss bushes and berries. (#3-#7)
Conclusions and Future Directions: There are some condition can affect the transect, like the components, weather and so on. By using the Hay infusion culture, we can observe protists and study different species of bacteria. This can help us to understand how natural selection leads the evolution of different species. AT
























