User:Camilla Dagum/Notebook/Biology 210 at AU: Difference between revisions
No edit summary |
No edit summary |
||
| (44 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
'''July 27 2014''' | |||
'''Introduction''' | |||
Several weeks ago bacteria from Transect 3 were cultured on agar and agar plus tetracycline plates. Samples of the colony growth had their 16S rRNA genes amplified via PCR, and these sequences were sent to a facility for direct sequencing (Bentley et al, 2014). Because of bacteria's tremendous diversity, we predict that there will be several different classes and genera of bacteria identified. | |||
'''Materials and Methods''' | |||
Samples of the colonies of bacteria growing on the agar and agar plus tetracycline plates were taken. The 16S rRNA gene from these samples was amplified using PCR and sent for direct sequencing. Once the sequences were returned, the ncbi BLAST tool was used. This website allows for input of nucleotide sequences and outputs gene matches. In this way the bacteria sampled are identified (Bentley et al, 2014). | |||
'''Results''' | |||
We analysed results not only from the samples we cultured, but also from other students' Transect 3 cultures. Only one of our three sampled cultures was able to be sequenced, and it turned out to be a ''Chryseobacteria'' culture. Three of the other five samples identified were also ''Chryseobacteria''. The other two samples were ''Flavobacteria'' and ''Aeromonas''. The summary of results is provided in Table 1. | |||
[[Image:Dagum_Bacteria_Table.jpg]] | |||
In the analysis of the bacteria done in an earlier lab, we found mostly gram-positive and rod-shaped bacteria. However, we may have been mistaken on gram-identification. A look at the images taken through the microscope of the stained bacteria indicate a pink color (July 9, 2014 Fig. 2, 4). At the time we identified any staining as gram-positive; however, we now realize that gram-negative bacteria stain pink (Bentley et al, 2014). | |||
'''Discussion''' | |||
Through sequencing we sought to identify the exact classes and genera of bacteria occupying Transect 3. We predicted that there would be a wide variety of bacteria found, but that hypothesis proved false. Four out of the six bacteria identified were ''Chryseobacteria''. This bacteria is abundant in soil environments (Kirby et al, 2004) so its dominance in the samples is understandable. Interestingly, the other two identified species don't seem to match our transect environment. These bacteria tend to be located in aquatic environments (Abell & Bowman, 2005) (Sartory). One explanation for this discrepancy is simply that even species with the same class can be quite different and occupy different niches. | |||
'''References''' | |||
1. Kirby, J., Sader, H., Walsh, T., & Jones, R. (2004). Antimicrobial Susceptibility and Epidemiology of a Worldwide Collection of ''Chryseobacterium'' spp: Report from the SENTRY Antimicrobial Surveillance Program (1997-2001). ''Journal of Clinical Microbiology''. Retrieved 2014 July 27 from http://jcm.asm.org/content/42/1/445.full.pdf | |||
2. Abell, GC., & Bowman, JP. (2005). Ecological and biogeographic relationships of class Flaveobacteria in the Southern Ocean. ''FEMS Microbiology Ecology''. Retrieved 2014 July 27 from http://www.ncbi.nlm.nih.gov/pubmed/16329875 | |||
3. Sartory, DP., et al. Aeromonas. ''WHO: Guidelines for Drinking Water Quality''. Retrieved 2014 July 27 from http://www.who.int/water_sanitation_health/dwq/admicrob2.pdf | |||
4. Bentley., Walters-Conte., & Zeller. (2014). ''Biology 210 Lab Manual''. Washington: American University. | |||
'''July 16 2014''' | |||
'''Introduction''' | |||
We have studied the bacteria, protists, plant life, and invertebrates that inhabit Transect 3. In this miniature lab, we turn the focus to vertebrates and ideas of ecological community within an ecosystem. Vertebrates are the most complex of the Earth's life forms, as the vertebrae and cranium protect the organism's central nervous system, allowing for more complex brains to develop. Vertebrates interact quickly and directly with their environment, giving them advantages over invertebrate organisms. Many vertebrates, such as the birds present in Transect 3, use invertebrates as a primary food source (Freeman et al, 2008). | |||
The interactions between the different domains and lineages of life within a geographic location are what form the ecosystem. One way to describe these interactions is through the use of a food web, which relates organisms by who feeds on whom. In this procedure, we will be creating a food web based on the organisms identified over the past three weeks that inhabit the Transect at various trophic levels. | |||
'''Materials and Methods''' | |||
Five vertebrates known or suspected to live in Transect 3 will be identified, and their ecological niches explored. The data collected on the bacteria, plant life, etc. that inhabit the Transect will be used in combination with the new information to compose a food web (Bentley et al, 2014). | |||
'''Results and Discussion''' | |||
The species of vertebrates likely to inhabit Transect 3 are shown in Table 1. | |||
[[Image:Dagum_Table_Vertebrates.jpg]] | |||
These are species common to environments similar to that of Transect 3. Blue jays, robins, and black-capped chickadees tend to eat invertebrates as well as tree seeds such as acorns in colder weather. They are common in residential areas and parks, especially those with shrubby bushes and trees (Blue Jay, 2011) (American Robin, 2011) (Black-capped Chickadee, 2011). These features are all characteristic of Transect 3, and so would benefit these birds. | |||
The eastern grey squirrel and eastern chipmunk are both also common to parks and residential areas with high density of bushes and trees. They tend to eat the flowers, seeds, and buds from different plants, as well as bird eggs (Lawniczak, 2002) (Kroll, 2013). These are all sources available within Transect 3. Additionally, chipmunks tend to burrow; they could certainly burrow into the soil in Transect 3. These animals also enjoy the nuts and seeds of trees not found in Transect 3, indicating that this transect is just one part of a much larger range. The bird species similarly have a range far greater than the one transect. | |||
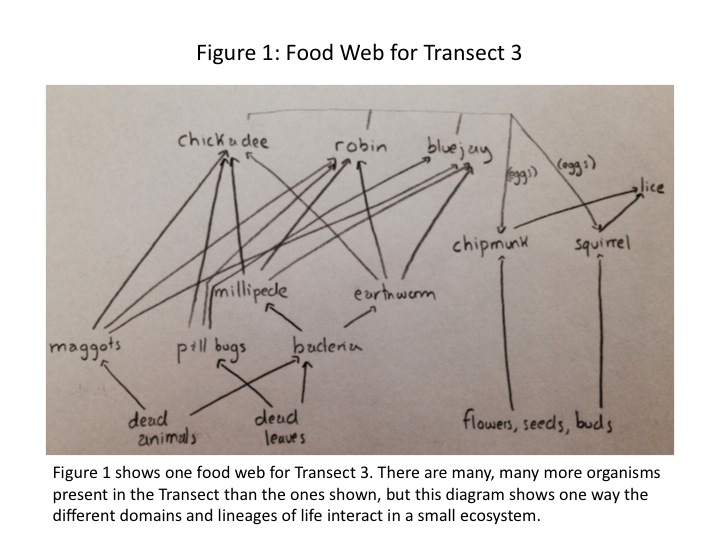
With all the main categories of life considered within the transect, we may create a possible food web that is occurring in the ecosystem. There are notably more species than we studied and identified over the course of the labs, so the food web shown in Fig. 1 is a small part of a much larger web of connections. | |||
[[Image:Dagum_Transect_3_Food_Web.jpg]] | |||
Ecological community is represented by these organisms by the network of ways in which they interact with each other and their environment. The bacteria, invertebrates, and vertebrates exist at different trophic levels, shown in Fig. 1. The birds are secondary or tertiary consumers, while the flower, seeds, and buds are from the primary producers. Carrying capacity of Transect 3 is the amount of life the transect can sustain. If the birds were limited to the resources in Transect 3, they would not be able to survive because the transect simply does not have the capacity to provide for many secondary or tertiary consumers. | |||
'''References''' | |||
1. 1. Freeman, S. et al. (2008). ''Biological Science'' (5th ed.). Boston, MA: Pearson. | |||
2. Bentley., Walters-Conte., & Zeller. (2014). ''Biology 210 Lab Manual''. Washington: American University. | |||
3. (2011). Blue Jay. ''All About Birds''. Ithaca, NY: Cornell Lab of Ornithology. Retrieved 2014 July 16 from http://www.allaboutbirds.org/guide/blue_jay/id | |||
4. (2011). American Robin. ''All About Birds''. Ithaca, NY: Cornell Lab of Ornithology. Retrieved 2014 July 16 from http://www.allaboutbirds.org/guide/American_Robin/id | |||
5. (2011). Black-capped Chickadee. ''All About Birds''. Ithaca, NY: Cornell Lab of Ornithology. Retrieved 2014 July 16 from http://www.allaboutbirds.org/guide/Black-capped_Chickadee/id | |||
6. Lawniczak, M. (2002). "Sciurus carolinensis". Animal Diversity Web. Retrieved 2014 July 16 from http://animaldiversity.ummz.umich.edu/accounts/Sciurus_carolinensis/ | |||
7. Kroll, M. (2013). "Tamias striatus". Animal Diversity Web. Retrieved 2014 July 16 from http://animaldiversity.ummz.umich.edu/accounts/Tamias_striatus/ | |||
'''July 15 2014''' | |||
'''Introduction''' | |||
Invertebrates are an incredibly diverse group, with fourteen different phyla of organisms. They also represent the most ancient animals in Earth's evolutionary history--porifera, the sponges, are the first animals. Sponges contain only two embryonic tissue layers, the ectoderm and the endoderm, and are radially symmetric (along with two other invertebrate phyla). A tremendous evolutionary leap occurred with the development of the mesoderm. The mesoderm gives rise to the circulatory system, muscle, and internal structures such as bone and organs. The three layers of embryonic tissue are present in the other invertebrate phyla, as well as the more complex animals (Freeman et al, 2008). | |||
In this lab we examine the soil invertebrates found in different transects. Most soil invertebrates are arthropods (Bentley et al, 2014), so we predict that the organisms we find will be primarily arthropods. Further, that there will be arthropods common to different transects, because of the range of environments common arthropods can tolerate. | |||
'''Materials and Methods''' | |||
Prepared cross-sections of ''Planaria'', nematode, and ''Annelida'' were studied under the microscope. A wet mount of live nematodes was prepared and their movement examined (Bentley et al, 2014). | |||
Invertebrate samples were retrieved from the Berlese funnels prepared in the last lab. The invertebrates found were placed in depression slides, examined under the microscope, and identified using a dichotomous key (Ward's, 1988). Descriptions of the organisms were also noted. | |||
'''Results''' | |||
Three types of worms were examined under the microscope. ''Planaria'' is an acoelomate, having no enclosed body cavity. Nematodes are pseudocoelomates, having an enclosed body cavity partially lined with mesoderm. They move with a thrashing motion. ''Annelida'' is a coelomate, with an enclosed body amity completely lined with mesoderm. Coelomates, such as earthworms, move with an undulating motion. The three cross-sections are shown in Fig. 1, and the live nematodes are shown in Fig. 2. | |||
[[Image:Dagum_Coelom_Variation.jpg]] | |||
[[Image:Dagum_Live_Nematodes.jpg]] | |||
The ethanol tube of the Berlese funnel did not have any invertebrates; however, three samples were found in the leaf litter remaining in the funnel. These Transect 3 samples were observed under the microscope and identified using a dichotomous key. A mallophaga, or biting louse, was found, as was a spider and a pill bug. The mallophaga and spider are shown in Figs. 3 and 4, respectively. We note the mallophaga's 6 legs, antennae, and hairy oval body. The spider is sharply segmented, with 8 legs and a large round thorax. The pillbug had a segmented exoskeleton, which allowed it to roll up into its characteristic ball shape. | |||
[[Image:Dagum_Biting_Lice_40x.jpg]] | |||
[[Image:Dagum_Spider_40x.jpg]] | |||
Transect 5 was also host to a mallophaga, with similar morphology to the Transect 3 mallophaga. From Transect 1 we identified a millipede, with its many-segmented body and numerous legs. The millipede is shown in Fig. 5. | |||
[[Image:Dagum_Millipede_40x.jpg]] | |||
All of our invertebrate finding and identifications are summarized in Table 1. | |||
[[Image:Dagum_Table_Invertebrates.jpg]] | |||
'''Discussion''' | |||
We studied several different phyla of invertebrates in this lab, ultimately identifying four different types of arthropods from the leaf litter samples of Transects 1, 3, and 5. We predicted that the leaf litter samples would primarily host arthropods, and that some arthropods are present in multiple transects. We found both of these predictions to be true. | |||
As shown in Table 1, the three samples from Transect 3-the mallophaga, spider, and pillbug-as well as the mallophaga from Transect 5 and the millipede from Transect 1 were all arthropods. They ranged in size from 1.1mm to 10mm, with the largest organism being the pillbug, and the smallest the lice. Further, both Transects 3 and 5 were host to biting lice. There were some morphological differences between the louse in Transect 3 and that in 5, however. The Transect 5 louse was larger and appeared to have a small tail-like protrusion at the end of its body that the Transect 3 sample did not have. There are more than 5,000 species of biting and sucking lice worldwide, however, so it is not surprising that the lice in the different transects were likely different species (Biting and Sucking Lice, 2014). | |||
Although no members of the phyla Platyhelminthes, Nematoda, or Annelida were observed from the Berlese funnels, we did have access to live samples and prepared cross-sections of these organisms. Their cross-sections are shown in Fig. 1. The earthworm (of phylum ''Annelida'') moves with an undulating motion, while the nematodes move with a thrashing motion. ''Annelida'' is the most complex of the phyla, having a fully-lined fluid-filled coelom (Bentley et al, 2014). This body structure allows it to have the most controlled and directional movement. Some platyhelminthes, the acoelomates, are unable to move at all. One example is the tapeworm (Freeman et al, 2008). | |||
Invertebrates compose a vital part of the Earth's ecosystems. Although only arthropods were found in the funnel products, we know that there are many different varieties of invertebrates present in the transect, because without these primary decomposers and consumer the ecosystem would fall apart. | |||
'''References''' | |||
1. Freeman, S. et al. (2008). ''Biological Science'' (5th ed.). Boston, MA: Pearson. | |||
2. Bentley., Walters-Conte., & Zeller. (2014). ''Biology 210 Lab Manual''. Washington: American University. | |||
3. (1988). ''A Simple Dichotomous Key to 18 Major Orders of Insects''. Rochester, NY: Ward's Natural Science. | |||
4. (2014). ''Biting and Sucking Lice''. London: The Amateur Entomologists Society. Retrieved 2014, July 15 from http://www.amentsoc.org/insects/fact-files/orders/phthiraptera.html | |||
'''July 13 2014''' | |||
'''Introduction''' | |||
Land plants first appear in fossils approximately 475 million years old, derived from the green algae that first appeared approximately 700 million years ago. The evolution of algae contributed to the rising oxygen levels in the Earth's atmosphere, ultimately leading to the Cambrian explosion. Extant algae and land plants are necessary for life on Earth, providing the oxygen needed by all aerobic respirators, as well as the chemical energy generated from sunlight. In terrestrial ecosystems, land plants are a major component and the dominant primary producers (Freeman et al, 2008). | |||
There are three major land plant categories: non-vascular plants, or bryophytes, seedless vascular plants, such as ferns, and seed plants. Seed plants can be further separated into angiosperms (flowering plants) and gymnosperms (non-flowering seed producers, such as pine trees). The seeds of angiosperms can be characterized as either monocot, having a single cotyledon, or dicot, having a pair of cotyledons. The various plant types have different reproductive methods and morphological characteristics. Non-vascular plants must be in water to reproduce, while the gametophytes of angiosperms, pollen, can be dispersed in air. Morphologically we find that non-vascular plants such as mosses are unable to grow to any significant height and lack specialized structures, while vascular plants have many specialized structures and can grow to tremendous heights (Freeman et al, 2008). | |||
Fungi are another immensely important kingdom of life. They are heterotrophic, and obtain food by secreting digestive enzymes and absorbing the digested matter. Fungi can be divided into three major divisions: the zygomycota, ascomycota, and basidiomycota. Zygomycota is distinguished by its formation of a zygospore for reproduction (Bentley et al, 2014). | |||
In this lab we examine five plant samples from Transect 3. Transect 3 is dominated by trees, bushes, and flowering plants, so we expect that the samples will have vascularization and specialized structures. | |||
'''Materials and Methods''' | |||
Five plant samples from around the transect were collected and brought back to the lab for analysis. The locations from where they were gathered were noted. The samples were compared to known samples in the lab to determine the types of plants gathered. The presence of vascularization, specialized structures, and how the plants were predicted to reproduce were noted. | |||
No fungi were collected, but samples of fungi were observed under the microscope. | |||
To prepare for the next lab, a Berlese funnel was set up. A 500g sample of surface litter from the transect was gathered. A piece of screening material was fitted and taped into the base of a funnel, and the funnel was placed on top of a tube containing approximately 25mL ethanol. The entire set-up is connected to a ring stand. To prevent ethanol evaporation, a strip of parafilm was wrapped around the connection between the funnel and the tube. The surface litter was then carefully transferred into the funnel, and the set-up moved under a light source. The lightbulb-funnel space is covered with foil, and the Berlese funnel is left to rest for five days (Bentley et al, 2014). | |||
'''Results''' | |||
Five plant samples were collected from various plants in the transect. The locations of each collection are shown in Fig. 1, and images of the plants from which they were taken are shown in Fig. 2. Each plant has many possibilities for sampling, so the individual samples are shown in Fig. 3. We took two flowers, both from flowering bushes, leaves from the two trees in the transect, and a leaf from one of the bushes. | |||
[[Image:Dagum_Transect_3_Plant_Samples_1.jpg]] | |||
[[Image:Dagum_Transect_3_Plant_Samples_3.jpg]] | |||
[[Image:Dagum_Transect_3_Plant_Samples_4.jpg]] | |||
These plants were all clearly vascular in nature. Depending on seed type, the vascular bundles would either be scattered (monocot seeds) or in a ring (dicot seeds). As no seeds were found in the transect, we cannot make absolute determination on vascular structure. However, the vascular structure of a lily plant was observed under the microscope, shown in Fig. 4. We note the scattered vascular bundles; this indicates a plant from a monocot seed. Those plants which we predict to be monocot will have a vascular structure similar to the image in Fig. 4. | |||
[[Image:Dagum_Lily_Stem_Cross_section.jpg]] | |||
Samples 1 and 5 were both flowers from the transect's many angiosperms. They are shown in greater detail in Fig. 5. We note some differences in flower structure; primarily that Sample 1 has many small flowers grouped together, whereas Sample 5 is from a plant which produces individual flowers. We also note the structures fundamental to all flowers, e.g. the pollen-covered anthers, the stigma and style, and the petals. | |||
[[Image:Dagum_Transect_3_Angiosperm_Samples.jpg]] | |||
The sample characteristics are summarized in Table 1. Seed-type was predicted based on morphological characteristics observed in each sample that are typical of either monocot or dicot seeds. | |||
[[Image:Dagum_Table_1_Plant_Samples.jpg]] | |||
The ''Rhizpus'' fungi observed under the microscope is shown in Fig. 6. We note the sporangia that produce the zygospores common to the zygomycota division of fungi. | |||
[[Image:Dagum_Rhizopus_Zygospores.jpg]] | |||
'''Discussion''' | |||
The purpose of this lab was to characterize the land plants growing in Transect 3. We predicted that the plants sampled would be vascular seed-producers, and that is what was found. As shown in Fig. 2, the transect plants were leafy bushes and trees. Only vascular seed-producing plants can grow to bushes and trees. The sizes of the leaves and flowers varied, from the small Sample 1 0.6cm flowers, to the larger Sample 5 5cm flowers. The Sample 4 leaf from one of the bushes was notably different from the tree leaves (Samples 2 and 3). It was much more waxy and stiff, and the bush leaf had a small spike on the end. The cluster arrangement of the leaves on all the sample plants was similar. They tended to feature a woodier stem or branch onto which the leaves attached. | |||
Once the plants were characterized as vascular seed-producers, we sought to determine what type of seeds were made for each sample. The summary of results is shown in Table 1. We determined whether a plant was monocot or dicot based on common characteristics. Monocot seed leaves tend to be longer and narrower, with a central vein. Leaves from dicot seeds tend to be broad, with a network of veins. As Fig. 3 shows, the Samples 2 and 4 have a single central vein, while Sample 3 is a broad leaf with a branching network of veins. For the flowers, we know that monocot flowers tend to have petals in multiples of three, while dicot flowers tend to have petals in multiples of four or five. Using this criterion, Samples 1 and 5 are both from dicot seeds (Bentley et al, 2014). | |||
This method of identification is not ideal. We note in Figs. 2 and 3 that Sample 1 has come from a plant with leaves that are longer and have a central vein. These features are more indicative of a monocot seed plant. If there was a way to obtain seeds from all the plants, that would be a superior analysis. | |||
Although no fungi were found in the Transect, we did observe a zygomycota fungus under the microscope, the ''Rhizopus''. This is shown in Fig. 6. We note the sporangia, which contain the zygospores. When the sporangia matures, it releases the spores, allowing for the spread of the ''Rhizopus'' fungus. It is also clear this is a fungi because of the filamentous structure. We can see the hyphae that have grown to form the sporangia. | |||
'''References''' | |||
1. Freeman, S. et al. (2008). ''Biological Science'' (5th ed.). Boston, MA: Pearson. | |||
2. Bentley., Walters-Conte., & Zeller. (2014). ''Biology 210 Lab Manual''. Washington: American University. | |||
'''July 9 2014''' | '''July 9 2014''' | ||
| Line 7: | Line 202: | ||
In this lab we will be studying the prokaryotic life present in the Hay infusion made from a soil sample of Transect 3. Different dilutions of the Hay infusion were grown on nutrient agar and tetracycline plates then analysed. It is likely that the colonies present are bacteria rather than archaea, as archaea tend to prefer extreme environments. Tetracycline is a common antibiotic that can affect both gram-positive and gram-negative bacteria by inhibiting protein synthesis. When a bacterial cell attempts to translate an mRNA, the tetracycline prevents binding of the aminoacyl tRNA to the A-site of the large ribosomal subunit. Resistance to tetracycline, however, is an increasingly common occurrence in many bacterial strains (Chopra et al, 2001). | In this lab we will be studying the prokaryotic life present in the Hay infusion made from a soil sample of Transect 3. Different dilutions of the Hay infusion were grown on nutrient agar and tetracycline plates then analysed. It is likely that the colonies present are bacteria rather than archaea, as archaea tend to prefer extreme environments. Tetracycline is a common antibiotic that can affect both gram-positive and gram-negative bacteria by inhibiting protein synthesis. When a bacterial cell attempts to translate an mRNA, the tetracycline prevents binding of the aminoacyl tRNA to the A-site of the large ribosomal subunit. Resistance to tetracycline, however, is an increasingly common occurrence in many bacterial strains (Chopra et al, 2001). | ||
We predict | We predict there will be a greater variety and number of bacteria that grow on the agar-only plates. If this is true, we expect that samples of colonies taken from agar plates will contain different bacteria, and those colonies that grow in the presence of tetracycline will tend to be more similar as all the bacteria must be resistant. | ||
'''Materials and Methods''' | '''Materials and Methods''' | ||
| Line 32: | Line 227: | ||
[[Image:Dagum_Gram_Stained_10_5_Agar.jpg]] | [[Image:Dagum_Gram_Stained_10_5_Agar.jpg]] | ||
[[Image:Dagum_Table_2_Bacteria_Characterization.jpg]] | [[Image:Dagum_Table_2_Bacteria_Characterization.jpg]] | ||
The Hay Infusion from which this bacteria was grown did not appear to have changed in character from the previous week. The colour and cloudiness was still similar, and there were no green shoots or obvious signs of life at the surface. | |||
'''Discussion''' | '''Discussion''' | ||
| Line 37: | Line 234: | ||
In this lab we sought to characterize the different bacterial species present in the Hay infusion culture of Transect 3, as well as compare the species that grew on nutrient agar plates with those that were able to grow in the presence of tetracycline. We predicted that there would be a greater variety in bacterial species on the nutrient agar plates, as well as a greater number of colonies. Both predictions held. | In this lab we sought to characterize the different bacterial species present in the Hay infusion culture of Transect 3, as well as compare the species that grew on nutrient agar plates with those that were able to grow in the presence of tetracycline. We predicted that there would be a greater variety in bacterial species on the nutrient agar plates, as well as a greater number of colonies. Both predictions held. | ||
As shown in Table 1, there were significantly more bacteria growing on the nutrient agar plates, with one of the tetracycline plates completely void of colonies. | As shown in Table 1, there were significantly more bacteria growing on the nutrient agar plates, with one of the tetracycline plates completely void of colonies. The difference in colony number between the agar and tetracycline plates is possibly even greater than was found in this experiment. We note that there is an unusually high number of colonies on the <math>10^{-9}</math> tetracycline plate, especially compared to plates with greater concentrations of Hay infusion. This is likely due to a contamination error made in plating the bacteria. The nutrient agar plates were all spread with the same spreader, inoculated in order of lowest to highest concentration--there was no need to clean the spreader between uses, as the concentration was increasing. However, we went from the <math>10^{-3}</math> agar plate to the <math>10^{-9}</math> tetracycline plate with the same spreader, neglecting to sterilize it. Thus we believe there is an inaccurate concentration of colonies on the apparently least-concentrated tetracycline plate. | ||
In Fig. 1 we note the greater variety of colonies present on the nutrient agar plates as compared to those growing in tetracycline--all the tetracycline colonies were uniform in colour and shape, and most were of the same size. The colonies growing on the agar, however, were of many different colours and shapes. | |||
Further differences between the colonies on the agar were found upon gram-staining inspection. As noted in Table 2, both gram-positive and gram-negative bacteria grew on the agar plates. Unfortunately as we only took one sample from the tetracycline colonies, we cannot be sure if gram-positive bacteria were more or less likely to grow in its presence. Although many antibiotics affect the bacterial cell wall, making them more detrimental to gram-positive bacteria, tetracycline affects protein synthesis (Chopra et al, 2001). Thus we cannot make a prediction about which bacteria type are more likely to be resistant. | |||
'''References''' | '''References''' | ||
Latest revision as of 13:29, 28 July 2014
July 27 2014
Introduction
Several weeks ago bacteria from Transect 3 were cultured on agar and agar plus tetracycline plates. Samples of the colony growth had their 16S rRNA genes amplified via PCR, and these sequences were sent to a facility for direct sequencing (Bentley et al, 2014). Because of bacteria's tremendous diversity, we predict that there will be several different classes and genera of bacteria identified.
Materials and Methods
Samples of the colonies of bacteria growing on the agar and agar plus tetracycline plates were taken. The 16S rRNA gene from these samples was amplified using PCR and sent for direct sequencing. Once the sequences were returned, the ncbi BLAST tool was used. This website allows for input of nucleotide sequences and outputs gene matches. In this way the bacteria sampled are identified (Bentley et al, 2014).
Results
We analysed results not only from the samples we cultured, but also from other students' Transect 3 cultures. Only one of our three sampled cultures was able to be sequenced, and it turned out to be a Chryseobacteria culture. Three of the other five samples identified were also Chryseobacteria. The other two samples were Flavobacteria and Aeromonas. The summary of results is provided in Table 1.
In the analysis of the bacteria done in an earlier lab, we found mostly gram-positive and rod-shaped bacteria. However, we may have been mistaken on gram-identification. A look at the images taken through the microscope of the stained bacteria indicate a pink color (July 9, 2014 Fig. 2, 4). At the time we identified any staining as gram-positive; however, we now realize that gram-negative bacteria stain pink (Bentley et al, 2014).
Discussion
Through sequencing we sought to identify the exact classes and genera of bacteria occupying Transect 3. We predicted that there would be a wide variety of bacteria found, but that hypothesis proved false. Four out of the six bacteria identified were Chryseobacteria. This bacteria is abundant in soil environments (Kirby et al, 2004) so its dominance in the samples is understandable. Interestingly, the other two identified species don't seem to match our transect environment. These bacteria tend to be located in aquatic environments (Abell & Bowman, 2005) (Sartory). One explanation for this discrepancy is simply that even species with the same class can be quite different and occupy different niches.
References
1. Kirby, J., Sader, H., Walsh, T., & Jones, R. (2004). Antimicrobial Susceptibility and Epidemiology of a Worldwide Collection of Chryseobacterium spp: Report from the SENTRY Antimicrobial Surveillance Program (1997-2001). Journal of Clinical Microbiology. Retrieved 2014 July 27 from http://jcm.asm.org/content/42/1/445.full.pdf
2. Abell, GC., & Bowman, JP. (2005). Ecological and biogeographic relationships of class Flaveobacteria in the Southern Ocean. FEMS Microbiology Ecology. Retrieved 2014 July 27 from http://www.ncbi.nlm.nih.gov/pubmed/16329875
3. Sartory, DP., et al. Aeromonas. WHO: Guidelines for Drinking Water Quality. Retrieved 2014 July 27 from http://www.who.int/water_sanitation_health/dwq/admicrob2.pdf
4. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
July 16 2014
Introduction
We have studied the bacteria, protists, plant life, and invertebrates that inhabit Transect 3. In this miniature lab, we turn the focus to vertebrates and ideas of ecological community within an ecosystem. Vertebrates are the most complex of the Earth's life forms, as the vertebrae and cranium protect the organism's central nervous system, allowing for more complex brains to develop. Vertebrates interact quickly and directly with their environment, giving them advantages over invertebrate organisms. Many vertebrates, such as the birds present in Transect 3, use invertebrates as a primary food source (Freeman et al, 2008).
The interactions between the different domains and lineages of life within a geographic location are what form the ecosystem. One way to describe these interactions is through the use of a food web, which relates organisms by who feeds on whom. In this procedure, we will be creating a food web based on the organisms identified over the past three weeks that inhabit the Transect at various trophic levels.
Materials and Methods
Five vertebrates known or suspected to live in Transect 3 will be identified, and their ecological niches explored. The data collected on the bacteria, plant life, etc. that inhabit the Transect will be used in combination with the new information to compose a food web (Bentley et al, 2014).
Results and Discussion
The species of vertebrates likely to inhabit Transect 3 are shown in Table 1.
These are species common to environments similar to that of Transect 3. Blue jays, robins, and black-capped chickadees tend to eat invertebrates as well as tree seeds such as acorns in colder weather. They are common in residential areas and parks, especially those with shrubby bushes and trees (Blue Jay, 2011) (American Robin, 2011) (Black-capped Chickadee, 2011). These features are all characteristic of Transect 3, and so would benefit these birds.
The eastern grey squirrel and eastern chipmunk are both also common to parks and residential areas with high density of bushes and trees. They tend to eat the flowers, seeds, and buds from different plants, as well as bird eggs (Lawniczak, 2002) (Kroll, 2013). These are all sources available within Transect 3. Additionally, chipmunks tend to burrow; they could certainly burrow into the soil in Transect 3. These animals also enjoy the nuts and seeds of trees not found in Transect 3, indicating that this transect is just one part of a much larger range. The bird species similarly have a range far greater than the one transect.
With all the main categories of life considered within the transect, we may create a possible food web that is occurring in the ecosystem. There are notably more species than we studied and identified over the course of the labs, so the food web shown in Fig. 1 is a small part of a much larger web of connections.
Ecological community is represented by these organisms by the network of ways in which they interact with each other and their environment. The bacteria, invertebrates, and vertebrates exist at different trophic levels, shown in Fig. 1. The birds are secondary or tertiary consumers, while the flower, seeds, and buds are from the primary producers. Carrying capacity of Transect 3 is the amount of life the transect can sustain. If the birds were limited to the resources in Transect 3, they would not be able to survive because the transect simply does not have the capacity to provide for many secondary or tertiary consumers.
References
1. 1. Freeman, S. et al. (2008). Biological Science (5th ed.). Boston, MA: Pearson.
2. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
3. (2011). Blue Jay. All About Birds. Ithaca, NY: Cornell Lab of Ornithology. Retrieved 2014 July 16 from http://www.allaboutbirds.org/guide/blue_jay/id
4. (2011). American Robin. All About Birds. Ithaca, NY: Cornell Lab of Ornithology. Retrieved 2014 July 16 from http://www.allaboutbirds.org/guide/American_Robin/id
5. (2011). Black-capped Chickadee. All About Birds. Ithaca, NY: Cornell Lab of Ornithology. Retrieved 2014 July 16 from http://www.allaboutbirds.org/guide/Black-capped_Chickadee/id
6. Lawniczak, M. (2002). "Sciurus carolinensis". Animal Diversity Web. Retrieved 2014 July 16 from http://animaldiversity.ummz.umich.edu/accounts/Sciurus_carolinensis/
7. Kroll, M. (2013). "Tamias striatus". Animal Diversity Web. Retrieved 2014 July 16 from http://animaldiversity.ummz.umich.edu/accounts/Tamias_striatus/
July 15 2014
Introduction
Invertebrates are an incredibly diverse group, with fourteen different phyla of organisms. They also represent the most ancient animals in Earth's evolutionary history--porifera, the sponges, are the first animals. Sponges contain only two embryonic tissue layers, the ectoderm and the endoderm, and are radially symmetric (along with two other invertebrate phyla). A tremendous evolutionary leap occurred with the development of the mesoderm. The mesoderm gives rise to the circulatory system, muscle, and internal structures such as bone and organs. The three layers of embryonic tissue are present in the other invertebrate phyla, as well as the more complex animals (Freeman et al, 2008).
In this lab we examine the soil invertebrates found in different transects. Most soil invertebrates are arthropods (Bentley et al, 2014), so we predict that the organisms we find will be primarily arthropods. Further, that there will be arthropods common to different transects, because of the range of environments common arthropods can tolerate.
Materials and Methods
Prepared cross-sections of Planaria, nematode, and Annelida were studied under the microscope. A wet mount of live nematodes was prepared and their movement examined (Bentley et al, 2014).
Invertebrate samples were retrieved from the Berlese funnels prepared in the last lab. The invertebrates found were placed in depression slides, examined under the microscope, and identified using a dichotomous key (Ward's, 1988). Descriptions of the organisms were also noted.
Results
Three types of worms were examined under the microscope. Planaria is an acoelomate, having no enclosed body cavity. Nematodes are pseudocoelomates, having an enclosed body cavity partially lined with mesoderm. They move with a thrashing motion. Annelida is a coelomate, with an enclosed body amity completely lined with mesoderm. Coelomates, such as earthworms, move with an undulating motion. The three cross-sections are shown in Fig. 1, and the live nematodes are shown in Fig. 2.
The ethanol tube of the Berlese funnel did not have any invertebrates; however, three samples were found in the leaf litter remaining in the funnel. These Transect 3 samples were observed under the microscope and identified using a dichotomous key. A mallophaga, or biting louse, was found, as was a spider and a pill bug. The mallophaga and spider are shown in Figs. 3 and 4, respectively. We note the mallophaga's 6 legs, antennae, and hairy oval body. The spider is sharply segmented, with 8 legs and a large round thorax. The pillbug had a segmented exoskeleton, which allowed it to roll up into its characteristic ball shape.
Transect 5 was also host to a mallophaga, with similar morphology to the Transect 3 mallophaga. From Transect 1 we identified a millipede, with its many-segmented body and numerous legs. The millipede is shown in Fig. 5.
All of our invertebrate finding and identifications are summarized in Table 1.
Discussion
We studied several different phyla of invertebrates in this lab, ultimately identifying four different types of arthropods from the leaf litter samples of Transects 1, 3, and 5. We predicted that the leaf litter samples would primarily host arthropods, and that some arthropods are present in multiple transects. We found both of these predictions to be true.
As shown in Table 1, the three samples from Transect 3-the mallophaga, spider, and pillbug-as well as the mallophaga from Transect 5 and the millipede from Transect 1 were all arthropods. They ranged in size from 1.1mm to 10mm, with the largest organism being the pillbug, and the smallest the lice. Further, both Transects 3 and 5 were host to biting lice. There were some morphological differences between the louse in Transect 3 and that in 5, however. The Transect 5 louse was larger and appeared to have a small tail-like protrusion at the end of its body that the Transect 3 sample did not have. There are more than 5,000 species of biting and sucking lice worldwide, however, so it is not surprising that the lice in the different transects were likely different species (Biting and Sucking Lice, 2014).
Although no members of the phyla Platyhelminthes, Nematoda, or Annelida were observed from the Berlese funnels, we did have access to live samples and prepared cross-sections of these organisms. Their cross-sections are shown in Fig. 1. The earthworm (of phylum Annelida) moves with an undulating motion, while the nematodes move with a thrashing motion. Annelida is the most complex of the phyla, having a fully-lined fluid-filled coelom (Bentley et al, 2014). This body structure allows it to have the most controlled and directional movement. Some platyhelminthes, the acoelomates, are unable to move at all. One example is the tapeworm (Freeman et al, 2008).
Invertebrates compose a vital part of the Earth's ecosystems. Although only arthropods were found in the funnel products, we know that there are many different varieties of invertebrates present in the transect, because without these primary decomposers and consumer the ecosystem would fall apart.
References
1. Freeman, S. et al. (2008). Biological Science (5th ed.). Boston, MA: Pearson.
2. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
3. (1988). A Simple Dichotomous Key to 18 Major Orders of Insects. Rochester, NY: Ward's Natural Science.
4. (2014). Biting and Sucking Lice. London: The Amateur Entomologists Society. Retrieved 2014, July 15 from http://www.amentsoc.org/insects/fact-files/orders/phthiraptera.html
July 13 2014
Introduction
Land plants first appear in fossils approximately 475 million years old, derived from the green algae that first appeared approximately 700 million years ago. The evolution of algae contributed to the rising oxygen levels in the Earth's atmosphere, ultimately leading to the Cambrian explosion. Extant algae and land plants are necessary for life on Earth, providing the oxygen needed by all aerobic respirators, as well as the chemical energy generated from sunlight. In terrestrial ecosystems, land plants are a major component and the dominant primary producers (Freeman et al, 2008).
There are three major land plant categories: non-vascular plants, or bryophytes, seedless vascular plants, such as ferns, and seed plants. Seed plants can be further separated into angiosperms (flowering plants) and gymnosperms (non-flowering seed producers, such as pine trees). The seeds of angiosperms can be characterized as either monocot, having a single cotyledon, or dicot, having a pair of cotyledons. The various plant types have different reproductive methods and morphological characteristics. Non-vascular plants must be in water to reproduce, while the gametophytes of angiosperms, pollen, can be dispersed in air. Morphologically we find that non-vascular plants such as mosses are unable to grow to any significant height and lack specialized structures, while vascular plants have many specialized structures and can grow to tremendous heights (Freeman et al, 2008).
Fungi are another immensely important kingdom of life. They are heterotrophic, and obtain food by secreting digestive enzymes and absorbing the digested matter. Fungi can be divided into three major divisions: the zygomycota, ascomycota, and basidiomycota. Zygomycota is distinguished by its formation of a zygospore for reproduction (Bentley et al, 2014).
In this lab we examine five plant samples from Transect 3. Transect 3 is dominated by trees, bushes, and flowering plants, so we expect that the samples will have vascularization and specialized structures.
Materials and Methods
Five plant samples from around the transect were collected and brought back to the lab for analysis. The locations from where they were gathered were noted. The samples were compared to known samples in the lab to determine the types of plants gathered. The presence of vascularization, specialized structures, and how the plants were predicted to reproduce were noted.
No fungi were collected, but samples of fungi were observed under the microscope.
To prepare for the next lab, a Berlese funnel was set up. A 500g sample of surface litter from the transect was gathered. A piece of screening material was fitted and taped into the base of a funnel, and the funnel was placed on top of a tube containing approximately 25mL ethanol. The entire set-up is connected to a ring stand. To prevent ethanol evaporation, a strip of parafilm was wrapped around the connection between the funnel and the tube. The surface litter was then carefully transferred into the funnel, and the set-up moved under a light source. The lightbulb-funnel space is covered with foil, and the Berlese funnel is left to rest for five days (Bentley et al, 2014).
Results
Five plant samples were collected from various plants in the transect. The locations of each collection are shown in Fig. 1, and images of the plants from which they were taken are shown in Fig. 2. Each plant has many possibilities for sampling, so the individual samples are shown in Fig. 3. We took two flowers, both from flowering bushes, leaves from the two trees in the transect, and a leaf from one of the bushes.
These plants were all clearly vascular in nature. Depending on seed type, the vascular bundles would either be scattered (monocot seeds) or in a ring (dicot seeds). As no seeds were found in the transect, we cannot make absolute determination on vascular structure. However, the vascular structure of a lily plant was observed under the microscope, shown in Fig. 4. We note the scattered vascular bundles; this indicates a plant from a monocot seed. Those plants which we predict to be monocot will have a vascular structure similar to the image in Fig. 4.
Samples 1 and 5 were both flowers from the transect's many angiosperms. They are shown in greater detail in Fig. 5. We note some differences in flower structure; primarily that Sample 1 has many small flowers grouped together, whereas Sample 5 is from a plant which produces individual flowers. We also note the structures fundamental to all flowers, e.g. the pollen-covered anthers, the stigma and style, and the petals.
The sample characteristics are summarized in Table 1. Seed-type was predicted based on morphological characteristics observed in each sample that are typical of either monocot or dicot seeds.
The Rhizpus fungi observed under the microscope is shown in Fig. 6. We note the sporangia that produce the zygospores common to the zygomycota division of fungi.
Discussion
The purpose of this lab was to characterize the land plants growing in Transect 3. We predicted that the plants sampled would be vascular seed-producers, and that is what was found. As shown in Fig. 2, the transect plants were leafy bushes and trees. Only vascular seed-producing plants can grow to bushes and trees. The sizes of the leaves and flowers varied, from the small Sample 1 0.6cm flowers, to the larger Sample 5 5cm flowers. The Sample 4 leaf from one of the bushes was notably different from the tree leaves (Samples 2 and 3). It was much more waxy and stiff, and the bush leaf had a small spike on the end. The cluster arrangement of the leaves on all the sample plants was similar. They tended to feature a woodier stem or branch onto which the leaves attached.
Once the plants were characterized as vascular seed-producers, we sought to determine what type of seeds were made for each sample. The summary of results is shown in Table 1. We determined whether a plant was monocot or dicot based on common characteristics. Monocot seed leaves tend to be longer and narrower, with a central vein. Leaves from dicot seeds tend to be broad, with a network of veins. As Fig. 3 shows, the Samples 2 and 4 have a single central vein, while Sample 3 is a broad leaf with a branching network of veins. For the flowers, we know that monocot flowers tend to have petals in multiples of three, while dicot flowers tend to have petals in multiples of four or five. Using this criterion, Samples 1 and 5 are both from dicot seeds (Bentley et al, 2014).
This method of identification is not ideal. We note in Figs. 2 and 3 that Sample 1 has come from a plant with leaves that are longer and have a central vein. These features are more indicative of a monocot seed plant. If there was a way to obtain seeds from all the plants, that would be a superior analysis.
Although no fungi were found in the Transect, we did observe a zygomycota fungus under the microscope, the Rhizopus. This is shown in Fig. 6. We note the sporangia, which contain the zygospores. When the sporangia matures, it releases the spores, allowing for the spread of the Rhizopus fungus. It is also clear this is a fungi because of the filamentous structure. We can see the hyphae that have grown to form the sporangia.
References
1. Freeman, S. et al. (2008). Biological Science (5th ed.). Boston, MA: Pearson.
2. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
July 9 2014
Introduction
The domain bacteria is one of the three domains of life. Bacteria are prokaryotes, single-celled organisms that lack complex membrane-bound organelles. Archaea is a second prokaryotic domain of life, and eukarya the third domain. Bacterial and archaeal lineages are the most ancient on Earth; the oldest fossils found, dating back 3.5 billion years ago, are carbon-rich deposits derived from bacteria. Eukaryotic life did not arise until 1.75 billion years ago. Bacteria are fantastically abundant and can proliferate in almost any environment. It is estimated that there are approximately [math]\displaystyle{ 5x10^{30} }[/math] individual bacteria and archaea alive today (Freeman et al, 2008).
In this lab we will be studying the prokaryotic life present in the Hay infusion made from a soil sample of Transect 3. Different dilutions of the Hay infusion were grown on nutrient agar and tetracycline plates then analysed. It is likely that the colonies present are bacteria rather than archaea, as archaea tend to prefer extreme environments. Tetracycline is a common antibiotic that can affect both gram-positive and gram-negative bacteria by inhibiting protein synthesis. When a bacterial cell attempts to translate an mRNA, the tetracycline prevents binding of the aminoacyl tRNA to the A-site of the large ribosomal subunit. Resistance to tetracycline, however, is an increasingly common occurrence in many bacterial strains (Chopra et al, 2001).
We predict there will be a greater variety and number of bacteria that grow on the agar-only plates. If this is true, we expect that samples of colonies taken from agar plates will contain different bacteria, and those colonies that grow in the presence of tetracycline will tend to be more similar as all the bacteria must be resistant.
Materials and Methods
The growth on the agar and agar plus tetracycline plates was observed with the naked eye. The number of colonies on each plate were counted, and the morphologies of the individual colonies were compared. Three colonies were chosen for further study: one from a tetracycline plate and two from different agar plates. Wet mounts of these colonies were prepared and observed under the microscope using the oil-immersion lens (1000x total magnification).
These same three colonies were then sampled to be gram-stained. To perform a gram stain, a sample of the colony is taken using a sterilized metal loop and smeared onto a slide. The location of the smear is noted by a wax pencil outline made on the underside of the slide. The slide is then passed through a flame three times to be heat fixed. Next, working above a staining tray, the smear is covered with crystal violet for 1 minute then rinsed off with water. The smear is then covered with Gram's iodine for one minute and again rinsed off, but with 95% alcohol. Once rinsed, the smear is covered with safranin stain for 20-30 seconds, and gently rinsed with water. The excess water is carefully blotted off with a paper towel. The sample is now gram-stained and ready to be observed under a microscope.
The three colonies were sampled for a third time to undergo a PCR reaction. A sample from each colony was transferred to a tube with [math]\displaystyle{ 100 \mu L }[/math] of water and incubated at [math]\displaystyle{ 100^oC }[/math] for ten minutes. The boiled samples are then centrifuged for five minutes at 13,400rpm. While the samples are centrifuging, [math]\displaystyle{ 20 \mu L }[/math] of primer mixture is added to each PCR tube with a PCR bead. [math]\displaystyle{ 5 \mu L }[/math] of the supernatant from the centrifuged samples is then transferred to the PCR tubes, and the tubes are placed in a PCR machine. The results of the PCR will be analysed in the following lab (Bentley et al, 2014).
Results
All the colonies on the tetracycline plates were yellow in colour and circular in shape with even margins. Those growing on the agar plates had considerably greater diversity, with colonies of different colours and shapes present. On the [math]\displaystyle{ 10^{-5} }[/math] agar plate there was a rhizoid lawn, and on the [math]\displaystyle{ 10^{-9} }[/math] agar plate one of the colonies was a deep red. Additionally, there were significantly more colonies growing on the plates without tetracycline. The colonies counted on each plate and the colonies/mL concentration are shown in Table 1. Figure 1 shows the individual plates, arranged by dilution level and presence of tetracycline.
The colonies chosen for further investigation were from the [math]\displaystyle{ 10^{-9} }[/math] tetracycline plate, the [math]\displaystyle{ 10^{-9} }[/math] nutrient agar plate, and the [math]\displaystyle{ 10^{-5} }[/math] nutrient agar plate. Description of each colony can be found in Table 2. The colonies were between 0.4 and 0.5cm, all convex, but different in colour and shape. Wet mounts of unstained bacteria samples did not yield much information, as the bacteria was difficult to discern. At 1000x magnification, all three bacteria samples looked identical, simply numerous black dots.
Once the bacteria were gram-stained, more useful data were taken. It was found that the [math]\displaystyle{ 10^{-9} }[/math] tetracycline sample and the [math]\displaystyle{ 10^{-5} }[/math] nutrient agar sample were both gram-positive, and the [math]\displaystyle{ 10^{-9} }[/math] nutrient agar sample gram-negative. Although the shapes of the individual bacteria were still difficult to discern, it appeared as though the [math]\displaystyle{ 10^{-9} }[/math] tetracycline sample was bacillus-shaped and the [math]\displaystyle{ 10^{-5} }[/math] nutrient agar sample coccus-shaped. Images of the stained bacteria at 100x magnification are shown in Figs. 2-5, and a summary of results in Table 2. None of the bacteria appeared to be motile.
The Hay Infusion from which this bacteria was grown did not appear to have changed in character from the previous week. The colour and cloudiness was still similar, and there were no green shoots or obvious signs of life at the surface.
Discussion
In this lab we sought to characterize the different bacterial species present in the Hay infusion culture of Transect 3, as well as compare the species that grew on nutrient agar plates with those that were able to grow in the presence of tetracycline. We predicted that there would be a greater variety in bacterial species on the nutrient agar plates, as well as a greater number of colonies. Both predictions held.
As shown in Table 1, there were significantly more bacteria growing on the nutrient agar plates, with one of the tetracycline plates completely void of colonies. The difference in colony number between the agar and tetracycline plates is possibly even greater than was found in this experiment. We note that there is an unusually high number of colonies on the [math]\displaystyle{ 10^{-9} }[/math] tetracycline plate, especially compared to plates with greater concentrations of Hay infusion. This is likely due to a contamination error made in plating the bacteria. The nutrient agar plates were all spread with the same spreader, inoculated in order of lowest to highest concentration--there was no need to clean the spreader between uses, as the concentration was increasing. However, we went from the [math]\displaystyle{ 10^{-3} }[/math] agar plate to the [math]\displaystyle{ 10^{-9} }[/math] tetracycline plate with the same spreader, neglecting to sterilize it. Thus we believe there is an inaccurate concentration of colonies on the apparently least-concentrated tetracycline plate.
In Fig. 1 we note the greater variety of colonies present on the nutrient agar plates as compared to those growing in tetracycline--all the tetracycline colonies were uniform in colour and shape, and most were of the same size. The colonies growing on the agar, however, were of many different colours and shapes.
Further differences between the colonies on the agar were found upon gram-staining inspection. As noted in Table 2, both gram-positive and gram-negative bacteria grew on the agar plates. Unfortunately as we only took one sample from the tetracycline colonies, we cannot be sure if gram-positive bacteria were more or less likely to grow in its presence. Although many antibiotics affect the bacterial cell wall, making them more detrimental to gram-positive bacteria, tetracycline affects protein synthesis (Chopra et al, 2001). Thus we cannot make a prediction about which bacteria type are more likely to be resistant.
References
1. Freeman, S. et al. (2008). Biological Science (5th ed.). Boston, MA: Pearson.
2. Chopra, I. & Roberts, M. (2001). Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiology and Molecular Biology Reviews, 65, 232-260. Retrieved July 9, 2014, from http://mmbr.asm.org/content/65/2/232.long
3. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
July 7 2014
Introduction
There are three domains into which all life is organized: archaea, bacteria, and eukarya. Archaea and bacteria are prokaryotes, lacking complex membrane-bound organelles. All prokaryotes are unicellular. Eukaryotes, however, can range in complexity from single-celled organisms to multi-celled plants, animals, and fungi. All multicellular organisms are eukaryotic (Freeman et al, 2008). In this lab we will be studying unicellular eukaryotes. Two large categories of such organisms exist; these categories are the algae, which are capable of photosynthesis, and the protists, which are heterotrophs (Bentley et al, 2014).
As described in the previous notebook entry, in the last lab we made a Hay infusion culture from a soil sample from Transect 3. The unicellular eukaryotes that will be identified are those that come from the soil sample Hay infusion. The Hay infusion has developed over the course of two days. We predict that within the Hay infusion there will be different ecosystems characterized by different organisms, based on the depth beneath the surface. If this is true, then different species will be identified at different layers of the infusion. We further predict that algae will likely be found exclusively in top areas of the Hay infusion.
Materials and Methods
The Hay infusion was carefully transported to the lab station, upon which time it was visually inspected. It is important to disturb the infusion as little as possible, as the infusion ecosystem changes with depth. Shaking the solution will disturb the protists and algae.
Two samples were taken from the infusion, one from the very top of the solution and one from the region near the bottom of the container. Two wet mounts were made from the samples, and each was studied under a compound microscope. Identification of two distinct unicellular eukaryotes from each region sampled was done using a dichotomous key, for a total of four organisms identified (Bentley et al, 2014).
After removing samples from the Hay infusion, a serial dilution of the solution was performed. The Hay infusion was mixed and a 100 [math]\displaystyle{ \mu L }[/math] sample taken. The 100 [math]\displaystyle{ \mu L }[/math] of Hay infusion was then added to 10mL of nutrient broth. This mixture, a 1:100, or [math]\displaystyle{ 10^{-2} }[/math] dilution was swirled to combine. 100 [math]\displaystyle{ \mu L }[/math] of the [math]\displaystyle{ 10^{-2} }[/math] dilution was then taken and added to 10mL of nutrient broth. This second mixture is now a 1:10000, or [math]\displaystyle{ 10^{-4} }[/math] dilution. The process is repeated two more times to make [math]\displaystyle{ 10^{-6} }[/math] and [math]\displaystyle{ 10^{-8} }[/math] dilutions (Bentley et al, 2014). A diagram of the serial dilution procedure is shown below.
After making the different dilutions, they were plated onto agar and tetracycline plates. To plate the dilutions, 100[math]\displaystyle{ \mu L }[/math] of solution was transferred to the agar plate and spread. This process was repeated for the four agar plates (one plate per dilution). For the tetracycline plates, the method was identical: 100[math]\displaystyle{ \mu L }[/math] of solution on each plate. The plates were capped and stored in the lab to incubate at room temperature for five days (Bentley et al, 2014).
Results
The Hay infusion was cloudy, with a yellow-green tinge. No green shoots or apparent life were seen at the surface of the infusion. The fibrous plant matter from the soil sample (wood chips and small sticks) had settled to the bottom of the infusion.
In the first sample, from the surface of the Hay infusion, an algae and a protist were identified. The algae was a deep blue-tinted green, and appeared to be composed of multiple cells. It was approximately 50[math]\displaystyle{ \mu m }[/math] across. The protist identified was a pair of Blepharisma, each approximately 25 [math]\displaystyle{ \mu m }[/math] across. The Blepharisma was a brownish pink and had cilia. It is shown in Fig. 1.
In the second sample, from the bottom of the Hay infusion, two distinct protists were identified. One organism identified was a Colpidium. The Colpidium was colourless, covered in cilia, and was quite small. It was approximately 12.5 [math]\displaystyle{ \mu m }[/math] across, and oval-shaped. Fig. 2 is a picture taken of the Colpidium through the microscope.
The second organism identified was a Chilomonas. It was approximately 37 [math]\displaystyle{ \mu m }[/math] across. It was an elongated cell with two locomotor flagella and was tinted green. We were unable to take a picture of the Chilomonas in the microscope; however, Fig. 3 is an image of Chilomonas taken from the Encyclopedia of Life (Feng et al).
All identifications were made using Ward's Natural Science dichotomous key for free-living protozoa (Ward's Natural Science, 2002).
Discussion
As predicted, different species of unicellular eukaryotes were found in the different layers of the Hay infusion observed. Algae was only found at the surface of the infusion. As algae are photosynthetic organisms, it stands to reason that they would be more successful in the sunnier environment. The protists observed at the bottom of the Hay infusion were different from those observed at the infusion's surface. This supports the initial prediction. The two regions of the Hay infusion had different qualities and components, and this was reflected in the protists that colonized the areas.
One of the organisms identified was a Blepharisma, shown in Fig. 1. This species, and the other organisms found, are qualified as life by meeting five fundamental characteristics. Blepharisma is a filter feeder, so it is able to acquire energy. It is made of cells, in this case, a single cell. It has a nucleus which stores genetic information, and although we did not see any Blepharisma actively dividing, they do reproduce. Finally, to be qualified as life an organism must be a product of evolution. We see similarities between Blepharisma and the other eukaryotes found in the infusion and documented by biologists; we know that Blepharisma is related to different species, thus it must be a product of evolution (Freeman et al, 2008).
It was difficult to locate the organisms under the microscope, which certainly influenced the results. Although the species found in the different layers observed were indeed different, so few organisms were found in the samples that it is possible some organisms were present in both layers. If the Hay infusion had been left to develop for a longer period of time, it is likely that we would have had more conclusive results.
If the Hay infusion had been observed for another two months, we expect to observe several changes. Initially, while the food source is still abundant, we expect a boom in protist population of many species. As time goes on, however, only some of those species will continue to proliferate, based on how well-adapted they are to the infusion environment. The protists that are more motile could be expected to thrive over those that are less motile, as they will be able to swim to different food sources as food grows scarce.
References
1. Freeman, S. et al. (2008). Biological Science (5th ed.). Boston, MA: Pearson.
2. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
3. (2002). Free-Living Protozoa. Rochester, NY: Ward's Natural Science. Retrieved July 7, 2014 from http://www.ocmboces.org/tfiles/folder1837/Free_Living_Protozoa%5B1%5D.pdf
4. Feng, A., Wie-song., & Patterson, D. Portrait of Chilomonas . Encyclopedia of Life. Retrieved July 7, 2014 from http://eol.org/data_objects/27474071
July 2 2014
Introduction
The ecosystems of an area as small as the American University campus are many and varied. One ecosystem present is an approximately 8mX6m transect of tall bushes. This region is characterized by several different types of bushes, both flowering and non-flowering, as well as some tall grasses and two different species of trees.
Materials and Methods
A macroscopical biological assay was performed simply by inspection of the transect. We documented the different varieties of plants present, noted their location as well as their individual characteristics.
A Hay infusion culture was also made using a sample of the soil, collected at the base of the Japanese maple tree. Approximately 10g of the soil sample was combined with 500mL filtered water and 0.1g of dried milk. The mixture was gently mixed until combined then left to develop in an uncapped jar for two days. The infusion was left undisturbed in the lab near a window (Bentley et al, 2014).
Results
An aerial-view diagram of the transect is shown in Fig. 1. Some plant varieties appeared in multiple locations of the transect, others only once. The two trees present were different species. The different colours are associated with a different plant, labeled accordingly. Fig. 2 shows actual images of the different plants found in the transect. There were abiotic components in the transect as well. A concrete sidewalk and four metal benches were present.
The transect was mostly flat, with some mounded soil where the plants were located. The soil was dry and composed primarily of wood chips. When collecting a sample for the Hay infusion, different insect species were found.
Discussion
The purpose of this lab was to characterize the macroscopic plant life in a transect of tall bushes. We predicted that there would be a number of different species present in even the small area (8mx6m) of the transect. As shown in Fig. 1, nine different species of plant life were found. Most of the plants were bushes of different heights and leaf shape; some were flowering and some were not. The different plant species are documented in images in Fig. 2. Interestingly, the conical flowers from one of the bushes are visually very similar to the flowers from the flowering tree. The morphology of the various bushes was similar as well.
Although the different species will all be competing for resources, it is actually beneficial to the ecosystem to have greater diversity. The results of a ten-year study performed by Dr.'s Tilman, Reich, and Knops showed that ecosystems with a greater number of plant species were more stable (Tilman et al, 2006). Transect 3 appeared to be a healthy ecosystem; none of the plants had dead branches or areas, and all were growing and flowering abundantly. The health of the ecosystem is certainly influenced by the variety of macroscopic plant life, and is likely influenced by the variety of microscopic life. In the following lab we will examine the unicellular life present in the soil of Transect 3.
References
1. Bentley., Walters-Conte., & Zeller. (2014). Biology 210 Lab Manual. Washington: American University.
2. Tilman, D., Reich, P., & Knops, J. (2006). Biosystem and Ecosystem Stability in a Decade Long Grassland Experiment. Nature, 441, 629-632. Retrieved July 6, 2014 from http://www.nature.com/nature/journal/v441/n7093/abs/nature04742.html