The BioBricks Foundation:Standards/Technical/E.coli promoter standard: Difference between revisions
| Line 4: | Line 4: | ||
[[Image:Proposed promoter standard.JPG|none|frame|600px|'''proposed standard A'''. green box identifies the 7bp sequence that would be required at the end of all standard BB promoters, the -10 box should be directly upstream of these 7 bp. n's are there to ensure standard spacing. BB junction in CAPS (it's not part of promoter).]] | [[Image:Proposed promoter standard.JPG|none|frame|600px|'''proposed standard A'''. green box identifies the 7bp sequence that would be required at the end of all standard BB promoters, the -10 box should be directly upstream of these 7 bp. n's are there to ensure standard spacing. BB junction in CAPS (it's not part of promoter).]] | ||
This basically follows from Chris & Reshma's discussion below. The only addition is that the standard requires a defined spacing between the -10 box and the CAT sequence. The A in the CAT sequence should be the transcription start site. The reasonable spacing along with the use of CAT (which is the 'consensus' -1,+1,+2 sequence - see [[:Image:Promoter table.JPG|table]]) should hopefully lead to predictable transcription start at the 'A'. Unfortunately the current Berkeley promoter library / R0040 don't conform to this standard. I suspect their transcriptional start is at the first A in the BB junction, but it's hard to know because the sequence isn't obviously optimal for a transcription start | This basically follows from Chris & Reshma's discussion below. The only addition is that the standard requires a defined spacing between the -10 box and the CAT sequence. The A in the CAT sequence should be the transcription start site. The reasonable spacing along with the use of CAT (which is the 'consensus' -1,+1,+2 sequence - see [[:Image:Promoter table.JPG|table]]) should hopefully lead to predictable transcription start at the 'A'. Unfortunately the current Berkeley promoter library / R0040 don't conform to this standard. I suspect their transcriptional start is at the first A in the BB junction, but it's hard to know because the sequence isn't obviously optimal for a transcription start though it's not bad (see fig). | ||
[[Image:Promoter table.JPG|thumb|left|300px|Table with nucleotide frequencies from 168 promoters<cite>McClure</cite>]] | [[Image:Promoter table.JPG|thumb|left|300px|Table with nucleotide frequencies from 168 promoters<cite>McClure</cite>]] | ||
[[Image:PromotersInReg.JPG|none|thumb|500px|'''berkeley / R0040 promoters''' have unknown transcription start sites (IMO). Bold sequence is where I suspect transcription start could be based on data from 112 known promoters<cite>McClure</cite>. The 1,2,4 are the number of promoters that had the same spacing and the same -1,+1 bases. (e.g. only 1 promoter had 4n's followed by gc where the c was the trans start. 2 had right spacing for ct and 4 had right spacing for ta). In contrast, 9 promoters had ca and same spacing as proposed standard. Underline is my best guess at start site. I underlined T in R0040 as that is the known +1 site for the wt (non-BB'd) promoter, however normally the T is an A, which is much more likely to be a start.]] | [[Image:PromotersInReg.JPG|none|thumb|500px|'''berkeley / R0040 promoters''' have unknown transcription start sites (IMO). Bold sequence is where I suspect transcription start could be based on data from 112 known promoters<cite>McClure</cite>. The 1,2,4 are the number of promoters that had the same spacing and the same -1,+1 bases. (e.g. only 1 promoter had 4n's followed by gc where the c was the trans start. 2 had right spacing for ct and 4 had right spacing for ta). In contrast, 9 promoters had ca and same spacing as proposed standard. Underline is my best guess at start site. I underlined T in R0040 as that is the known +1 site for the wt (non-BB'd) promoter, however normally the T is an A, which is much more likely to be a start.]] | ||
Revision as of 02:34, 30 March 2008
Jason R. Kelly 02:11, 30 March 2008 (EDT):Seems like we should just make a decision about where to locate transcription start site (+1 site) in BB promoters. There's excellent previous discussion on the topic here (I also copied a portion of it below).
A proposal
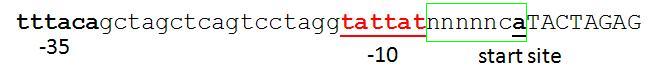
This basically follows from Chris & Reshma's discussion below. The only addition is that the standard requires a defined spacing between the -10 box and the CAT sequence. The A in the CAT sequence should be the transcription start site. The reasonable spacing along with the use of CAT (which is the 'consensus' -1,+1,+2 sequence - see table) should hopefully lead to predictable transcription start at the 'A'. Unfortunately the current Berkeley promoter library / R0040 don't conform to this standard. I suspect their transcriptional start is at the first A in the BB junction, but it's hard to know because the sequence isn't obviously optimal for a transcription start though it's not bad (see fig).


- Jason R. Kelly 04:59, 30 March 2008 (EDT):The other option is to adopt the R0040/Berkeley promoter set as the promoter standard. E.g. (-10box)nnnnnc(BBjunction). Downside here is that (1) I don't know how dependent the "non-consensus" transcriptional start will be on the n's (so might get different start sites w/ diff promoters) and (2) I don't know exactly where the transcriptional start is in the first place. Though I might just not be up to speed on best way to predict start site (ref below was best I could find). Of course this standard would have the advantage that we already have some parts in the format ;)

Previous discussion
JCAnderson: The site: http://parts.mit.edu/registry/index.php/Help:BioBrick_Prefix_and_Suffix under the BioBrick Prefix section has a really critical piece of information on how to design biobrick basic parts, and I think we should add to that a preferred way of biobricking the promoter initiation site relative to the polylinker to avoid heterogeneity 5' to the biobrick junction. Again, it is an arbitrary standard, and the options are (with the transcription start in bold): Define it like r0040(and what iGEM2006 did for the family of constitutive promoters):
- ...ctACTAGT
Or have it in the biobrick site explicitly, something like:
- ...ACTAGT
So that nothing has to be re-made, and so that more native promoter sequence can be present in the part I lean towards defining the standard as the r0040-compatible version.
Clearly not all promoters are going to be compatible with this standard. Some promoters have operators that overlap or extend beyond the transcriptional start. When making basic promoter parts, one has to currently make an arbitrary decision as to where to put the 3' end of the promoter. It would be preferrable to have a standard.
- Reshma 11:12, 21 August 2006 (EDT): I agree that we should have a default standard for the promoter-RBS junction. But in looking at the sequence logo for E. coli promoters, I think the typical nucleotides for the -1 and +1 positions are CA. In the absence of any strong reason to go with another scheme, why not go with E. coli promoter consensus? So perhaps something like ...
- ...caTACTAGAG
- i.e. Pretty similar to the R0040-compatible version but with the transcription start site being a defined nucleotide where possible along with the nucleotide before. It might make it a bit more likely that the transcription start site occurs where we think it should occur.
Reference
- Hawley DK and McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237-55. DOI:10.1093/nar/11.8.2237 |
- Horwitz MS and Loeb LA. DNA sequences of random origin as probes of Escherichia coli promoter architecture. J Biol Chem. 1988 Oct 15;263(29):14724-31.