Shlo/notebook/fall07: Difference between revisions
| Line 688: | Line 688: | ||
[[IMAGE: shlo121096wellgel2wanalyis.jpg]] | [[IMAGE: shlo121096wellgel2wanalyis.jpg]] | ||
=12/12= | |||
96 well plate made: using 1:100 diluted minipreps of controls | |||
*Mek in A-D | |||
**1ul in A and B | |||
**3uL in C | |||
**10uL in D | |||
*pdz in E-H | |||
**1ul in E and F | |||
**3uL in G | |||
**4uL in H | |||
Also, in parallel, ran "normal" E-gel, all with 3uL of miniprep. | |||
*Loaded: | |||
**Lane 1: Ladder | |||
**Lane 2-6: Pdz | |||
**7-12: mek | |||
Revision as of 13:01, 12 December 2007
Back to notebook
| Topocloning pdf | Topocloning Protocol | General Protocols| 1kb ladder| Low-range Egel Ladder
9/9
Grew up:
- RACE+AIDA-his(1)
- RACE+AIDA-strep
- RAE-strep2
- RAE-his3
- TS3
9/10 and 9/11
Overnight plate reader run.
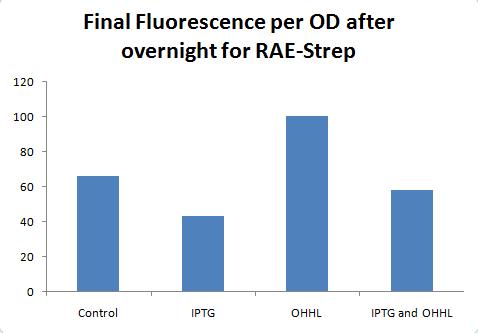
Used TS3 (columns 1-3), RAE-his3 (4-6), RAE-strep2 (7-9). Row B = straight dilution, C = w/ IPTG, D = w/OHHL, E= w/ IPTG & OHHL
More results to be posted soon.
Results for RAE-tagged are not that great, but maybe I'm just not used to the small range (not a huge difference between induced and non-induced).
9/12
Using only TS3 today because of yesterday's results.
Induced w/ different concentrations of IPTG, incubate at 37C. Will split into two aliquots each: one to be washed pre-addition of OHHL, others left w/ all IPTG and then OHHL addition. Plate reader to be run overnight.
9/13
Induced with different amounts of IPTG (9 different concentrations), incubated for 2.5 hr, took ODs. Diluted to same OD, spun down pellets in microcentrifuge tube, put in box in my freezer.
Also, collected data from last night's overnight run.

"Washed" corresponds to a 3x wash with PBS before reconstitution in LB+Amp+Kan to an OD analogous to the "nonwashed" control sample.
9/14
Ran direct magnetic binding assay with senders and non-tagged const. GFP, testing for noninduced binding and the like. Results were not spectacular. Control plates indicate that noninduced senders have a growth advantage, but when induced, const. GFP has the growth advantage.
More data to come.
9/15
More plate reader - exploring washing after IPTG induction, detecting growth of fluorescence.
Also, another direct magnetic selection assay with tagged receivers (iptg-induced, washed, grown again, then selected for).
9/16
Results from Yesterday
Selection/targeting plates
6.8x enrichment:

Control corresponds to non-induced tagged sender with equal amount of induced, tagged receiver. (no selection, just plating). As expected, the noninduced sender seems to grow much better (about 10x).
Experimental corresponds to strep-tagged receiver (induced) with his-tagged sender (noninduced). Selection for receiver was performed using streptactin beads. Compared to control, 6.8x enrichment.
Not shown: I tried strep-receiver (induced) with strep-tagged sender (noninduced), with streptactin beads. Since senders were noninduced, I still expected enrichment of receiver. Instead, I got both senders and receivers, and at a ratio analogous to the control. This indicates that the senders may be sticky even without induction; however, due to the experimental result above, I suspect the stickiness (leakiness of expression) still lends itself to specificity (eg, the noninduced his-sender does not stick to streptactin, but noninduced strep-sender does). My next assay will involve testing for enrichment capabilities *without* induction at all since IPTG seems so intrusive on fluorescence expression.
Plate Reader
Results show that washed cells display fluorescence growth significantly higher than non-washed cells, but in longer run, noninduced cells still show 2x as much absolute fluorescence. Perhaps more washes / growth before OHHL induction will allow for more fluorescence growth from washed samples. The two graphs below are adjusted for initial GFP values (equilibrated between samples). Graphs are also available with samples starting from the same OD (but not same GFP, apparently; perhaps washing [and almost definitely induction] negatively affects this). These will be posted soon.
9/17
Started another plate reader (re-do from summer work, exploring ratios of sender vs receiver).
Also, another plate selection assay to see whether noninduced cells stick/are enriched enough to be used without induction.
9/18
Yesterday's Plate Reader Results
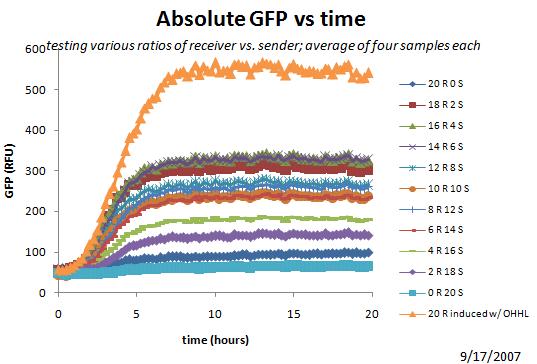
Above shows that the optimal ratio for eventual GFP may be around 15 receiver, 5 sender.
9/20
Growing up some cultures: will start with nickel plates tomorrow.
9/22
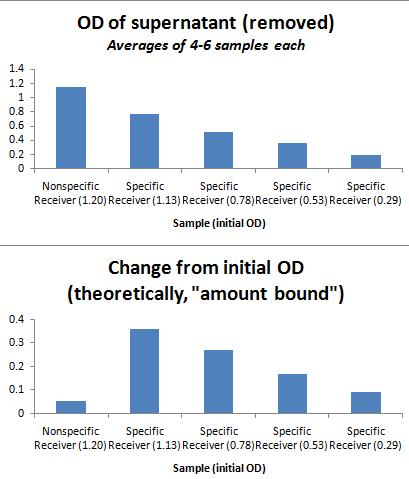
Nickel Plate Results (all averages of 4-6 wells each). Assay involved measuring ODs on both the photospectrometer and the plate reader, binding in a 37C shaking plate reader over 30 minutes. Afterward, the supernatant was removed from each well, and the well was refilled with LB. These results are shown below.
As shown in the graph below, the addition of equal ODs of His-sender (specific) and Strep-sender (nonspecific) results in a greater final OD left on the Nickel-plate for the specifically-bound senders.

Additional parts of assay included washing with imidazole 3x (10 min shaking, per wash). The clear plate was again refilled with LB. The final OD after washes was extremely low for all samples, with no distinction between specific and nonspecific binding; I suspect that the washing was too vigorous (or the imidazole, at 50mM, too high in concentration), and all specifically bound cells were washed away as well, leaving only small residues. This is a step that may be improved, should I attempt to repeat this experiment for greater enrichment. I also wonder if it is worth repeating this experiment, allowing for binding in a longer time frame (perhaps 1 hr instead of 30 minutes), which may also increase enrichment.
Additional results (added 9/24)
9/25
Redoing selection assay, except with receivers. Need to figure out control for the addition of sender.
Preliminary Graphs
9/27
The following shows "dilutions" of senders (actually, added in various amounts of other non-RFP cells). It fits a linear eq pretty well:


The image below shows that the washed wells showed no drastic increases in fluorescence, as the synthetic-OHHL-treated nonwashed wells did. This indicates that I may be washing away so many receiver cells that no fluorescence is detected, or I am not waiting long enough to detect substantial fluorescence increases (... I would have to start with a larger OD in my kinetic run, since the plates are almost all drying out already by the end of the 24 hr assay). In my re-do assay, perhaps I should not wash with imidazole at all, or allow the cells to bind for longer (though these already bound for 1.5 hr) before washes.

9/30
Redoing assay ... realized that I should bind senders first, then wash, then add receiver, since we probably need a lot less sender than receiver for detectable fluorescence.
I had washed w/ PBS (no imidazole) because I was afraid the imidazole was preventing all binding. This turned out to be a bad idea because I got a lot of nonspecific binding; that is to say, the strep binding was similar to the his. I did show that, with the amount of his-senders that were left in the well, I was able to activate the GFP expression for the receivers:
(These results are also sketchy because the plate reader was unplugged by someone in the lab accidentally, so I don't know how growth really went ..., and they weren't at 37 or shaking).
It's also worth noting that the cells were not washed after receiver was added, so I don't know how well the receivers are binding. I guess I want to make sure I get enrichment (almost NO strep ... or detectable difference in fluorescence growth curves, not available for this experiment) before I move onto that. That's the next step: bind senders, wash, bind receivers, wash, grow.
10/01
For today's new assay, I washed with imidazole, tested same things (see yesterday). Hopefully will get no GFP in the strep-sender lanes.
Results:
- I am unable to fully prevent binding of strep (nonspecific); GFP response is same for both strep and his - should be much higher for his.
- Need to do more washes in future?
10/06
- More washes done for longer
- Results still inconclusive / bad.
- Fluorescence on this clear plate doesn't seem to be going that well.
10/06-10/16 assays
Unsuccessful plate reader assays. Plate reader sent away for repair.
10/16-10/17 selection assays
- RFP + AIDAP1+2 (antiserum, 10uL): considerably fewer colonies than normal beads. 66x4 (all apparently white?)
- (4) control: AIDA-strep-send + JsLO (1) = 4 x (82 red, 235 white)
- DB JsLo (2) + Strep, 10ul: 24 red, 292 white
- 3 control: AIDA-strep + Aida p(1) = 8 x (63 red, 81 white)
- AIDa-his send + aida pdz 1+2 (10) = 4 x (65 red, 96 white)
- 2 control: 1:1000 dil, 1ul: 41 red, (126x2) white
- 3 strep- RFP + Jslo1 (1:100 dil) 10ul: 9 red, 12 white
- 4 strep RF+Jslo1 (1:100 dil 1ul): 3 red, 2 white
- 5 strep-RFP _ OmpP2 1:100 dil 10 ul: 22 red, 36 white
- 6 strep-RFP OmpP2 1:100 dil 1 ul: 5 red 1 white
Control sample with preserum antibodies shows a lot fewer colonies, as expected
10/18-10/19 selection
Grew up 10/18; IPTG-induce 8:30 am 10/19
- 1, strep+JsLoB (no pdz), normal bead, 5uL: 22 white, 4 red
- 1, 50uL: 120 white, 97 red
- 2, strep + JsLo Pdz-1, 5uL: 13 red, 0 white
- 2, 50uL: 67 red, 7 white
- 3, strep + jslo pdz-2, 5uL: 10 red, 2 white
- 3, 50uL: 51 white, 80 red
- 4, strep + pdz (1+2) 5uL: 2 white, 8 red
- 4, 50uL: 55 white, 78 red
- 5, strep + nonind pdz (1) 5uL: 5 white, 5 red
- 5, 50uL: 38 white, 78 red
- 5 control (~0.1 ul): 86 white, 4 red
- 6: strep + non-induced pdz (1+2), 5uL: 2 white, 5 red
- 7: strep + pdz(1+2) + PS bead: 11 red, 0 white
10/21-10/22 Selection
3 washes in MACS column, 4 washes with direct bead assay
(original dilution: 200 white, 600 red - to equilibriate OD)
last wash w/ 2x imidazole
- 1: JsLo pdz 1 + strep (10uL: 13 red, 0 white)
- 50uL: 90 red, 9 white
- 2: JsLo pdz 2 + strep (10uL 18 red, 0 white)
- 50uL: 92 red, 11 white
- CONTROL (eq of 0.01uL, preselection): 8 x (84 white, 79 red)
- 3: JsLo Pdz 1+2, + strep RFP (10uL: 21 red, 2 white)
- 50uL: 102 red, 1 white
- CONTROL: 33 white: 7 red ratio
10/22-10/23 Selection
Washes w/ 2x imidazole
- AIDA-1+strepR (4) control, eq to 0.01 uL: 130 red, 135 white
- 4, 10uL: 50 colonies, all white
- 4, 50uL: 15 red, 272 white
- 5: AIDA 2 + strepR, 10uL: 16 red, 492 white
- 6: AIDA 1+ 2+ strepR, 10uL: 12 red, 90 white
10/28
Designed Primers (credit to Perry for tons of help):
OmpA1_F
5' GAAATAATTTTGTTTAACTTTAAGAAGGAGatatacatatgaaagctactaaa 3'
OmpA1mek2_R_1
5' GGGTGTGCCGGGCTG GCTAGCGTTGTCCGGACGAGT 3'
OmpA1mek2_R_2
5' GGAAGCTTCTGCAGTTATCACACGGCGGTGCGCGTGGGTGTGCCGGGCTGGCT 3'
First round of PCR OmpA1_F/OmpA1mek2_R_1 at 58dC.
Using product from first round of PCR, second round of PCR OmpA1_F/OmpA1mek2_R_2 at 58dC.
10/31
- Electroporation of T9002 and S23I07 into Pdz 1, 2, 1+2 (6 combinations total)
- 1st pcr for Mek, using 5 ng OmpA_1-His (his tag should be removed by end of pcr).
11/1
- 2nd pcr
- colony pcr setup
11/2
- ran colony pcr gel
- pcr purification on mek 2nd pcr product
11/3 - 11/4
iGEM Jamboree!
11/5
- Running new colony PCR of AIDA-pdz's and S23I or T02.
- Did a Topo-ligation / top10 transformation for the Omp1 + Mek.
11/6
- Colony PCR plate grew, which is good news enough. Waiting for the actual PCR to finish so I can run an E-gel.
- PCR machine on other side of bench
- transformation of OmpA1+Mek actually yielded colonies; about 150 on the 50uL plate, and proportionally higher on the 250uL plate. The colonies are quite small though. Need to run colony PCR on them ...
- In PCR-1.
- Grew up JSLo-1, 2, 1+2, and B ... and J04450 (kan). Perhaps another selection assay soon just to calm my nerves about the JSLos.
- Colony PCR: Cotransformations
11/8
Things I need to do:
- Grow up successful Mek-tagged bacteria. Miniprep, digest, extract vector from LppOmpA1 and insert and ligate/transform into BL21 (DE3)s.
- Cotransform (via electroporation) with S23I and T02.
- Grow up successful Mek-tagged bacteria (doesn't matter whether they're cotransformed yet, but should be in BL21(DE3), obvi). Do magnetic bead assay with Prot-A coated beads, Lpp-OmpA-pdz (cotransformed, so that fluorescent proteins distinguish the bacteria).
- Grow up successful cotransformed bacteria for magnetic assays - perhaps one without Mek, just to ensure the pdz domains still are functional, and then with Mek to see whether the cells will aggregate.
- Use microscope to visualize said Mek-pdz interactions.
11/9
Mini-prepped Mek, digest of OmpAinPet29B and Mek w/ Hind III and NdeI. Left in pcr machine in the next bench over.
11/10
For some reason my clonewell only gives me the vector and not the Mek (500bp) insert. I'm going to redo the Mek col PCR and Mek miniprep, but continue with the extraction of vector ... (colony PCR is machine opposite my bench)
Also, growing up some cultures to co-transform quorum parts into JsLos (why did I transform them into AIDA ...? not sure).
- Lane 1: 1kb ladder
- Rest: Colony PCRs of Mek
11/12
- Cotransformed JsLo w/ SI and SE (6 samples total); good time constants
- Growing up JsLo for more selection assays (tomorrow?)
- Growing up Mek for miniprep/digest (tomorrow?)
11/13
- JsLO IPTG induction @ 8:15am
- Mek miniprep
- JsLO cotransformation colony PCR, should be done circa 11am
- JsLO cotransformation gel
- Lanes
- 1: 1 + SE
- 2: 2 + SE
- 3: 1+2 + SE
- 4: 1 + SI
- 5: 1 + SI
- 6: 2+ SI
- 7: 1+2+SI
- 8: Ladder
- 9: 1+2+SI
- 10: 1+2 + SE
- 11: 1+2 + SE
- 12: 2 + SE
11/15
Interestingly, of my 4 Mek digests (2, 3, 5, 7), only 7 seems to be giving me Clonewell bands - but Clonewell was performed.
Speed Vac'd, ligation w/ Roche quick ligation. Put in freezer for transformation ... soon.
11/17
Transformation day.
- ligation nanodrop: 126ng/uL
- 2uL into 50uL cells
- incubation started 9:50 am
11/19
Mek colony PCR for BL21-DE3's.
(all worked - yay!)
11/20
Electroporation (cotransformation) of Mek's with SI and JT. Wanted to do a third with T02 but can't find miniprep. (should make more myself)
Controls for selection assay didn't grow up - I might have to throw away that (really old) master plate and make a new one. Started growing up new controls when I got to lab; not sure they'll be grown enough for an assay this afternoon.
IPTG-induced Mek and pdz samples.
Performed selection assay, grew up T02 and S23I (nontagged) for new master plate and miniprep.
11/21
Moved plates into fridge because I needed to leave for Thanksgiving - unable to count colonies. Also, set aside some T02/S23I for miniprep and master-plating (in fridge - should hopefully be okay when I return on ... Sunday? ... to tend to them).
11/26
Colony counts and miniprep of SI, T02, and I22. Cotransformations for later this week?
Also, microscopy today of "mixed colonies" from selection assay.
Microscopy
- Clumping of pdz-tagged bacteria around ProtA-pdz beads
RFP microscopy didn't turn out too well ..
My images are generally disappointing. I'll have to run these again.
Other notes
Colony notes:
- Sample 1, with bead and Mek alone, had a few residual colonies but much fewer (maybe roughly 1/5 the amount?) compared to selection plates.
- Sample 2: bead, OmpA1-Mek and pdz-SI: looks like it has "mixed" colonies on selection plate! Wonder where I can take this ...?
- Controls had too many colonies; hard to compare any results with anything.
Miniprep (ng/uL):
- SI: 157.4
- T02: 331.1
- I22: 220.8
11/28
Grew up 6 samples (E1-E6) of "mixed colonies" from selection plates 3 and appropriate controls. Miniprep, gel with OmpA1F, OmpA1_Mek_2_R, and Pdz_2_R primers.
Appearances of overnight cultures:
- E1: white, perhaps VERY FAINT pinkish tint?
- E2: pink
- E3: white
- E4: white
- E5: red
- E6: pink
- All controls looked white
11/29
- Lane 1: ladder
- 2, 3: White controls
- 4, 5: Red controls
- 6, 7: Mixed controls
- 8-12: Experimental
11/30
12/1
- w/BB primers
- Expect bands of size 1584bp ONLY on pdz bacteria. Mek bacteria do not have any biobrick parts in them.
Lanes:
- 1kb Ladder
- Experimental Mixed Colony (E)1
- E2
- E3
- E4
- E5
- E6
- Mek control (Miniprepped and PCR'd, from Mek only plate)
- Pdz control
- Pdz+Mek control (mixed culture pre-miniprep)
- E5 control (no PCR)
- E6 control (no PCR)
- w/pdz reverse primer and OmpA1 forward primer
Lanes:
- 1kb Ladder
- Experimental Mixed Colony (E)1
- E2
- E3
- E4
- E5
- E6
- Mek control (Miniprepped and PCR'd, from Mek only plate)
- Pdz control
- Pdz+Mek control (mixed culture pre-miniprep)
- pdz control (no PCR)
- Pdz*Mek control (no PCR)
12/03
Talked w/ Alain about what to do next (will do another selection assay, try to run a lot of PCRs in parallel to quantify selection/Mek-pdz interaction to some extent)
Grew up liquid cultures
12/04
Selection assay day (urgh)
Need to try new PCR-prep:
- Take a portion of a colony and resuspend it in 25 ul of water
- Heat to 95C for 6 min
- Freeze immediately at -80C for at least 10 min
- Set up PCR reaction using 6 ul of the heated/frozen template (finalvolume of the PCr reaction: 50 ul).
So did selection (20 plates - in 37dC incubator) and PCR-preps; in PCR machine near door.
12/05
Selection:
- 1 = pdz bead (20uL) + JSLopdz1+2 (white before assay ...?) IPTG +, SI nontagged (500 uL each)
- 2 = pdz bead + Mek 7 IPTG + SI nontagged
- 3 = pdz bead + JsLo non-IPTG, SI nontagged
- 4 = pdz bead + JsLo IPTG +, Mek +
- 5 = pdz bead + JsLO IPTG - (non-induced), Mek IPTG +
Results
- 1 = pdz bead (20uL) + JSLopdz1+2 (white before assay ...?) IPTG +, SI nontagged (500 uL each)
- 1:10000 control = 72 colonies
- 20uL = 10 colonies
- 200uL = 66 colonies (all white?)
- 2 = pdz bead + Mek 7 IPTG + SI nontagged
- 1:100 control: 71 colonies
- 1:10,000 control = 9 colonies
- 20 uL = 0 colonies
- 200 uL = 2 colonies (2 bubbles?)
- 3 = pdz bead + JsLo non-IPTG, SI nontagged
- 1:10000 control: 1/4 = 210
- 20 uL = 94 colonies (1/4)
- 4 = pdz bead + JsLo IPTG +, Mek +
- 1:10000 control : 1/8 = 135
- 5 uL selection: 13 colonies
- 20 uL selection: 31 colonies
- 200 uL: 1/4 = 235 colonies
- 5 = pdz bead + JsLO IPTG - (non-induced), Mek IPTG +
- 1:10000 dilution: 1/4 = 150 colonies
- 20 uL = 64 colonies
- 200 uL = 164 colonies = 1/4
Gel (PCR prep attempt
Lanes
- 1kb ladder
- JsLo - 7 PDZ1+2
- JsLO 7 sample 2
- JSLo - 8 PDZ1+2
- JsLo 8 sample 2
- Mek 7
- SI-NT (control)
- JsLo - 9 PDZ1+2
- JsLO 9 sample 2
- JsLO - 10 PDZ1+2
- JsLo 10 sample 2
- Mek 1
12/06
Used old PCR-preps to make PCR rxns with BB primers. Will run gel later today.
UGHHH.
- a samples have 1uL of prepped DNA
- b samples have 5uL of prepped DNA
- controls were not PCRed
- All are Pdz1+2 SI's
- 1kb ladder
- 1a
- 1b
- 2a
- 2b
- 4a
- 4b
- 5a
- 5b
- 6a
- 1 control
- 4 control
Streaked a bunch of samples from my latest selection plate: only PDZ for now ... Running a PCR on them; ready for a Gel tomorrow.
12/07
Ran out of E-gels, but running one (will run another tonight)
(Lane 1 = 1 kb ladder, Lanes 2-12: Samples 1-11 from Mek/Pdz (4) Mixed Plate)
12/08
- Row A: PDZ1+2 (from control plate), prepped first, PCR'd
- Row B: samples from Row A straight from plate to PCR
- Row C: Mek (from control plate), prepped first, PCR'd
- Row D: samples from Row C straight from plate to PCR
- Row E: 1-6: from selection plate, using PDZ primers, prepped first; 7-12: no colonies (supposedly negative control; to make sure contamination in prep did not occur)
- Row F: samples from Row E straight from plate to PCR
- Row G: Biobrick primers, on pdz1+2 S23I's (should see 1584 bp)
- Row H: S23I straight from control plate
12/09
40uL was used for each PCR reaction, using a master mix derived from: http://openwetware.org/wiki/IGEM:Harvard/2007/Protocols#PCR.
Interestingly after the PCR/thermocycler, the reactions again seemed to dry up. I wonder if there's something wrong with the PCR caps I'm using? Am I applying them incorrectly?
- Row A: 10uL PCR prep from yesterday's plate, row A (pdz controls)
- Row B: 5uL of above
- Row C: "regular" PCR, colony picked from yesterday's swab plate
- Row D: 10uL PCR prep from yesterday's plate, row C (Mek controls)
- Row E: 5uL of above
- Row F: colony picked from yesterday's swab plate
- Row G: 1-6: pdz, from old control plates; 8-12: Mek
- Row H: same as above
12/10
- A-D: pdz
- E-H: Mek
... Results poor again.
Re-running PCRs, this time using the same master to make "normal" reactions in the other machine ... I got positive results in the past, so I should get these again ...
Attempt 2
Normal Egel

From same master mix, using same colony/swab. Put in regular (larger/500uL) tubes, in separate PCR machine - with same program.
- Ladder
- pdz F12
- pdz E2
- pdz E5
- pdz G2
- pdz H7
- Mek A1
- Mek A6
- Mek B3
- Mek B9
- Mek D9
- pdz F5
96 well egel
Using previously confirmed plates: Mek's from A-D; pdz1+2's from E-H
12/12
96 well plate made: using 1:100 diluted minipreps of controls
- Mek in A-D
- 1ul in A and B
- 3uL in C
- 10uL in D
- pdz in E-H
- 1ul in E and F
- 3uL in G
- 4uL in H
Also, in parallel, ran "normal" E-gel, all with 3uL of miniprep.
- Loaded:
- Lane 1: Ladder
- Lane 2-6: Pdz
- 7-12: mek