Sbb14 Adrianna Carrasco: Difference between revisions
No edit summary |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 170: | Line 170: | ||
[[Image:gel2bands.jpg]] | [[Image:gel2bands.jpg]] | ||
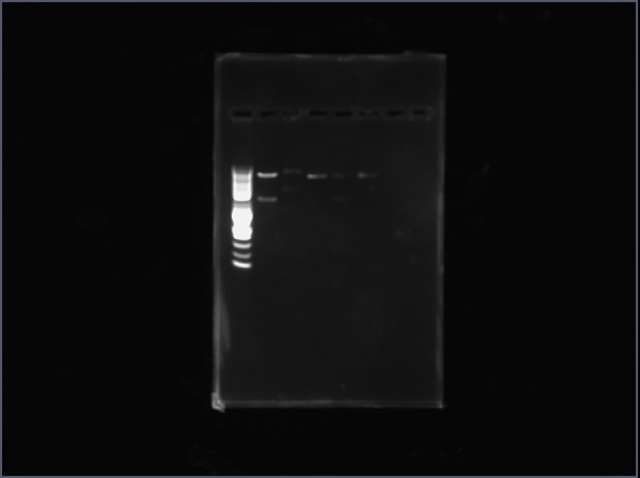
The column is the digested Met trna-synthetase and vector it was placed in. The fifth column is digested Asn trna-synthetase with the vector it was placed in. | The third column is the digested Met trna-synthetase and vector it was placed in. The fifth column is digested Asn trna-synthetase with the vector it was placed in. | ||
==Tuesday April 22== | |||
I sent my mini prep samples, Met 3 and Asn 1, to be sequenced. | |||
==Thursday April 24== | |||
I received my sequence analysis of each sample. Asn trna-synthetase was partial perfect and confirmed successful ligation into the plasmid vector. However, Met trna-synthetase had a bad read and did not match to any part of my sequence. This can be a result of a contaminated mini prep sample or ethanol residue. I will need to run another gel of another Met mini prep and check for appropriate band length then send it to be sequenced. | |||
Latest revision as of 12:32, 25 April 2014
Tuesday February 18
Found trna synthetase for Asn and Met in Thermus Thermophilus genome. Created following oligos for Asn and Met trna synthetase;
Forward asn-trna synthetase oligo: aggctCCATGGcacgcgtcttcattgacgagat
Reverse asn-trna synthetase oligo: ggtccGAATTCttaggggcgcatcctcgtgta
Methione had an internal Nco1 restriction site, and required a silent mutation.
Forward external Met trna-synthetase oligo 001: aggctCCATGGaaaaggtcttctacgtg
Reverse internal Met trna-synthetase oligo with silent mutation 003: ttgacgtaggccacggcctcctc
Forward internal Met trna-synthetase oligo with silent mutation 004: gaggaggccgtggcctacgtcaa
Reverse external Met trna-synthetase oligo 002: ggtccGAATTCttacttcaccaccgccccgg Adrianna Carrasco 13:38, 18 February 2014 (EST)
Thursday February 20
Today I created the construction file for Met trna-synthetase. I used the soeing protocol since there were ncor1, Bsmb1 and Bsa1 sites in the ORF.
Construction of Met trna-synthetase basic part
PCR Oligo Met 001/Oligos Met 003 on Therm genome (1242 bp, gp = A) PCR Oligos Met 004/Oligos Met 002 on Therm genome (656 bp, gp = B) --------------------------------------------------- PCR Oligos Met 001/Oligos Met 002 on A+B (1875 bp, Ncol/EcoR1) Digest pBAD/his A (NcoI/EcoR1, ?, L) Product is pBAD/his A-Met --------------------------------------------------- Oligos qbs 001 Forward Nco1 for Biobrick extreme variant of Met aggctCCATGGaaaaggtcttctacgtg Oligos qbs 003 Removing the Ncol site in Met trna-synthetase ttgacgtaggccacggcctcctc Oligos qbs 004 Removing the Nco1 site in Met trna-synthetase gaggaggccgtggcctacgtcaa Oligos qbs 002 Reverse EcoR1 for Met trna-synthetase ggtccGAATTCttacttcaccaccgccccgg
Adrianna Carrasco 13:35, 20 February 2014 (EST)
Thursday February 27
Today in lab, we watched the demo for the mini prep and gel electrophoresis. Then each group spilt up and performed the mini prep protocol for the plasmid.
Tuesday March 4
Today we set up our PCR reactions for our oligos. I had to do sowing for Methione, so that required extra internal oligos. We created a master mix buffer congaing ddH2O, buffer, and the DNTPs. We then diluted our oligos to 10μM and combined 'master mix' template DNA and our polymerase enzyme. Both PCR reactions were set in the 2k thermocycling since they were under 2kb.
Thursday March 6
My PCR reactions did not work, so I could not run the gel. I had to do PCR again for my SOeing MET and ASN oligos. They were both placed in the 2k thermocycling and the previous PCR protocol was used again.
Friday March 7
Today we ran a gel on our PCR products. My ASN PCR was of correct bp size at 1342bp. My first half of MET SOeing product did not work but the second half did. I did PCR again for the first SOeing product.
Tuesday March 11
Today we ran a gel on our PCR products. My first half of SOeing was successful. The expected bp range was around 1200bp. My two PCR bands are the second and third.
Thursday March 13
We did a zymo clean up on our successful PCR products. For ASN I followed the regular zymo protocol. For my MET products I followed the SOeing protocol and used double the amount of ADB buffer. After the zymo clean up for MET, I did the second PCR using my products from the first round as template DNA. I used the 2k55 thermocylcler.
Tuesday March 18
Today I ran a gel on my second PCR of my SOeing reaction for methionyl-trna synthetase. The gel had two distinct bands, one of which was the correct size of my expected product and another of smaller size. I cut out the correct size on the gel(1875bp). The smaller band could have been the oligos that I was joining together that may have been replicated a few times. After the correct band was cut out, I performed a zymo gel purification.
--Adrianna Carrasco 15:36, 18 March 2014 (EDT)
Thursday March 20
Digested my PCR gel purified product- Metdig and Asndig. Also digested our vector. The enzymes used to digest product and vector were NcoI and EcoRI.
The following protocol was used:
8uL of eluted PCR product
1uL of NEB Buffer 2 0.5uL EcoRI 0.5uL NcoI
Tuesday April 1
Today I ligated my vectdig and pcrdig (Metdig and Asndig). The following protocol was used:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase
We incubated for 30min on a bench top, then put on ice and continued onto the transformation process.
Transformation by heat shock: 1. Thaw a 200 uL aliquot of cells on ice
2. Add 50 uL of water to the cells (if greater volume is desired) 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (for Miniprep product, dilute by 10, then use 1uL of dilution) 5. Add 70 uL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 90 seconds at 42 (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour 10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight
Thursday April 3
Our transformation was successful and we had multiple white colonies. We then picked out the four best colonies from each plate and put them in LB media using the following protocol:
For each construct you will pick and later miniprep 2 colonies Add 4mL of LB media with the appropriate antibiotics to a clean test tube Pick a well-isolated, round, and "normal" looking colony with a toothpick Drop it in the test tube Incubate at 37 overnight
Tuesday April 8
Our selected colonies grew in the LB media. We did a mini prep of 2 selected colonies.
MIniprep purification of DNA
Pellet 2 mL saturated culture by spinning full speed, 30 seconds in a 2mL Microcentrifuge tube. Dump supernatant Add 250uL of P1 buffer into each tube. Resuspend the cells thoroughly Add 250uL of P2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down. Solution should become clearer. Add 350uL of N3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then shake. Spin in centrifuge at top speed for 5 minutes. Label blue columns with an alcohol-resistant lab pen. Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds. Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin) Wash each column with 500 uL of PB buffer. Spin in centrifuge at full speed for 15 seconds, then flick out the liquid again. Wash with 750uL of PE buffer (washes the salts off the resins). Spin in centrifuge at full speed for 15 seconds and flick out liquid again. Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol. Label new Microcentrifuge tubes and put columns in them. Elute them by adding 50uL of water down the middle of the column (don't let it stick to the sides). Spin in centrifuge at top speed for 30 seconds. Take out columns and cap the tubes. Clean up - note the P1 buffer is stored at 4degC and all the rest at room temperature.
Thursday April 10
We did our restriction mapping for our purified plasmids. We chose to digest with NcoI and EcoRI since the part inserted was about 2000bp long and would give a distinct band other than our vector. However, only one band was observed and we need to do another mini prep and gel.
Restriction mapping
7 uL ddH2O
1uL Miniprepped plasmid
1uL 10x NEB Buffer 2
0.5uL EcoRI
0.5uL BamHI
Tuesday April 15
Today we did a mini prep of our other two LB colonies. We followed the mini prep purification protocol for DNA.
Thursday April 17
We did restriction mapping for our purified plasmids. We digested with Ncol and EcoRl. There were two bands observed of correct size for Methione trna synthetase and Asparagine trna-synthetase. The expected size of the bands for Met-trna are 4053bp and 1879bp and for Asn-trna the expected bands are 4053bp and 1342bp.
The third column is the digested Met trna-synthetase and vector it was placed in. The fifth column is digested Asn trna-synthetase with the vector it was placed in.
Tuesday April 22
I sent my mini prep samples, Met 3 and Asn 1, to be sequenced.
Thursday April 24
I received my sequence analysis of each sample. Asn trna-synthetase was partial perfect and confirmed successful ligation into the plasmid vector. However, Met trna-synthetase had a bad read and did not match to any part of my sequence. This can be a result of a contaminated mini prep sample or ethanol residue. I will need to run another gel of another Met mini prep and check for appropriate band length then send it to be sequenced.