Sbb14-dbs: Difference between revisions
No edit summary |
No edit summary |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
==[[User:Daniel Steiner|Daniel Steiner]] 14:43, | ==[[User:Daniel Steiner|Daniel Steiner]] 14:03, 29 April 2014 (EDT)== | ||
Reverse read of SerS #4 confirmed the sequence as perfect. The read of ArgS #3 shows 170 bases of the beginning of the vector/gene (including ~20 of the gene), then a repeat of about the first 20bp of the gene, then 800bp of homology to the end of the gene (last 20bp) and vector. These two regions of the gene both contain CGCGC preceded by another ~5 C/G's. This points to error in insertion of the PCR product into the vector, possibly caused by the two ends of the gene annealing to each other at the similar regions. | |||
==[[User:Daniel Steiner|Daniel Steiner]] 13:34, 24 April 2014 (EDT)== | |||
Ran ArgS #1-4 between two gels, and none of the samples appeared to work. Submitted ArgS #3 for sequencing anyway, just to see if I possibly have product and just keep having errors with digestion. Also ordered reverse read of SerS #4 because it was perfect partial. | |||
[[Image:4_24_14_Gel_pdtvectdig.jpg]] | |||
Analytical Gel 1 order: Vectdig Trp1 Trp2 Tsquare TΔ Tx Arg1 Arg2 Arg3 Ladder | |||
[[Image:4_24_14_Gel2_pdtvectdig.jpg]] | |||
Analytical Gel 2 order: Trp3 Trp4 Tcircle Arg4 Vectdig Ladder | |||
==[[User:Daniel Steiner|Daniel Steiner]] 13:34, 22 April 2014 (EDT)== | |||
Submitted the miniprep of SerS #4 to Chris for sequencing. Did an analytical digestion of ArgS #1-4 with EcoRI/NcoI using 3μL of DNA instead of 1μL. | |||
==[[User:Daniel Steiner|Daniel Steiner]] 14:49, 17 April 2014 (EDT)== | |||
Digested the vector, ArgS #1-4, and SerS #1-3 with NcoI/EcoRI for restriction mapping. Ran a gel, and only the template plasmid gave more than one band. | |||
==[[User:Daniel Steiner|Daniel Steiner]] 14:43, 15 April 2014 (EDT)== | |||
Miniprepped 2mL of cell cocktail from the other four tubes of LB/ampicillin/transformed E. coli (ArgS #1/3, SerS #1/3). | Miniprepped 2mL of cell cocktail from the other four tubes of LB/ampicillin/transformed E. coli (ArgS #1/3, SerS #1/3). | ||
==[[User:Daniel Steiner|Daniel Steiner]] 14:43, | ==[[User:Daniel Steiner|Daniel Steiner]] 14:43, 10 April 2014 (EDT)== | ||
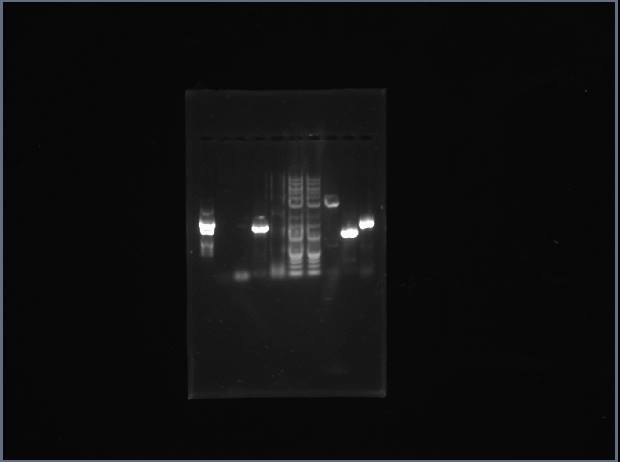
Did a 10μL analytical digest (1μL of DNA) of ArgS #2, ArgS #4, SerS #2, SerS #4, and the template plasmid (all with EcoRI and NcoI) for restriction mapping. Ran a gel, expecting to see two bands for the template plasmid ( | Did a 10μL analytical digest (1μL of DNA) of ArgS #2, ArgS #4, SerS #2, SerS #4, and the template plasmid (all with EcoRI and NcoI) for restriction mapping. Ran a gel, expecting to see two bands for the template plasmid (4055bp and 1252bp), two for ArgS (4055bp and 1781bp), and three for SerS (4055bp, 739bp, and 528bp). All bands were very faint, and only SerS #4 and the template plasmid gave correct bands. | ||
[[Image:4-10gel.jpg]] | [[Image:4-10gel.jpg]] | ||
The right gel (from left to right): | The right gel (from left to right): template, Adrianna's Met and Asn (2 lanes each), ArgS #2, ArgS #4, SerS #2, SerS #4, ladder. | ||
==[[User:Daniel Steiner|Daniel Steiner]] 15:05, 8 April 2014 (EDT)== | ==[[User:Daniel Steiner|Daniel Steiner]] 15:05, 8 April 2014 (EDT)== | ||
Latest revision as of 11:06, 29 April 2014
Daniel Steiner 14:03, 29 April 2014 (EDT)
Reverse read of SerS #4 confirmed the sequence as perfect. The read of ArgS #3 shows 170 bases of the beginning of the vector/gene (including ~20 of the gene), then a repeat of about the first 20bp of the gene, then 800bp of homology to the end of the gene (last 20bp) and vector. These two regions of the gene both contain CGCGC preceded by another ~5 C/G's. This points to error in insertion of the PCR product into the vector, possibly caused by the two ends of the gene annealing to each other at the similar regions.
Daniel Steiner 13:34, 24 April 2014 (EDT)
Ran ArgS #1-4 between two gels, and none of the samples appeared to work. Submitted ArgS #3 for sequencing anyway, just to see if I possibly have product and just keep having errors with digestion. Also ordered reverse read of SerS #4 because it was perfect partial.
Analytical Gel 1 order: Vectdig Trp1 Trp2 Tsquare TΔ Tx Arg1 Arg2 Arg3 Ladder
Analytical Gel 2 order: Trp3 Trp4 Tcircle Arg4 Vectdig Ladder
Daniel Steiner 13:34, 22 April 2014 (EDT)
Submitted the miniprep of SerS #4 to Chris for sequencing. Did an analytical digestion of ArgS #1-4 with EcoRI/NcoI using 3μL of DNA instead of 1μL.
Daniel Steiner 14:49, 17 April 2014 (EDT)
Digested the vector, ArgS #1-4, and SerS #1-3 with NcoI/EcoRI for restriction mapping. Ran a gel, and only the template plasmid gave more than one band.
Daniel Steiner 14:43, 15 April 2014 (EDT)
Miniprepped 2mL of cell cocktail from the other four tubes of LB/ampicillin/transformed E. coli (ArgS #1/3, SerS #1/3).
Daniel Steiner 14:43, 10 April 2014 (EDT)
Did a 10μL analytical digest (1μL of DNA) of ArgS #2, ArgS #4, SerS #2, SerS #4, and the template plasmid (all with EcoRI and NcoI) for restriction mapping. Ran a gel, expecting to see two bands for the template plasmid (4055bp and 1252bp), two for ArgS (4055bp and 1781bp), and three for SerS (4055bp, 739bp, and 528bp). All bands were very faint, and only SerS #4 and the template plasmid gave correct bands.
The right gel (from left to right): template, Adrianna's Met and Asn (2 lanes each), ArgS #2, ArgS #4, SerS #2, SerS #4, ladder.
Daniel Steiner 15:05, 8 April 2014 (EDT)
All 8 of the tubes of E. coli/LB/Amp had significant cell growth, so I miniprepped 2mL of the cell cocktail from each of tubes ArgS #2, ArgS #4, SerS #2, and SerS #4.
Daniel Steiner 15:56, 3 April 2014 (EDT)
Had cell growth on both plates, so I picked 4 isolated colonies from each plate and added each colony to a tube of 5mL LB broth + Ampicillin to incubate/shake overnight at 37°C.
Daniel Steiner 13:33, 1 April 2014 (EDT)
We checked our plates the day after plating and had no cell growth, so I re-ligated, transformed, and plated ArgS and SerS today.
Used the same protocol as last time, sans the recovery step (adding 2YT and incubating/shaking for 1 hour).
Daniel Steiner 16:42, 20 March 2014 (EDT)
Finished the Zymo Gel Purification of our vector digest.
Ligated the vector with our part digests (ArgS and SerS) and ran a negative control (vector without any insert).
Transformed E. coli with our plasmids using the following procedure (except we added 2YT without incubating/shaking afterward):
1. Thaw a 100 uL aliquot of cells on ice 2. IGNORE WATER ADDING STEP 3. Add 35μL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (for Miniprep product, dilute by 10, then use 1μL of dilution) 5. Add 65μL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 90 seconds at 42°C (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100μL of 2YT, let shake in the 37°C incubator for 1 hour 10. Plate 70+μL on selective antibiotics, let incubate at 37°C overnight
Plated my ArgS and SerS (along with Kevin's ThrS and ValS, Neeka's TrpS, and our negative control) on Ampicillin agar plates. We expect colonies on all of the plates, but much fewer on the negative control since those E. coli should not have functional plasmids with the Amp-R gene.
Daniel Steiner 15:42, 18 March 2014 (EDT)
Did a 30μL digest of our mini-prepped vector:
15μL vector, 3μL NEBuffer2, 1.5μL EcoRI, 1.5μL NcoI, 9μL ddH2O
Incubated for 1 hour @ 37°C.
Ran a gel to separate the fragments, cut out the larger band from each of the three wells, and melted them together in 600μL ADB buffer.
Daniel Steiner 16:23, 13 March 2014 (EDT)
Did regular Zymo Cleanup of Arg PCR product.
Digested the cleaned Ser PCR product with BsmBI (incubated in NEBuffer 3 at 55°C for 1 hour).
Digested the cleaned Arg PCR product with NcoI/EcoRI (incubated in NEBuffer 2 at 37°C for 1 hour).
Did regular Zymo Cleanup of both digests to get rid of the enzymes.
Daniel Steiner 16:16, 11 March 2014 (EDT)
We ran an analytic gel of our PCR products from last time (mine are two separate reaction tubes of Arg PCR product).
from left to right: Christy's Met, Adrianna's two Met pieces for soeing, Pro, ladder, Pro, two Arg, and two Thr.
Daniel Steiner 17:17, 7 March 2014 (EST)
We set up the PCR reactions again for our failed tRNA synthetases (P, Q, R, T), making two PCR tubes of each in case one fails. I set up the reaction for arginyl tRNA synthetase.
We also did a regular Zymo Cleanup of the products of the successful PCR reactions (S, V, W, and Y). I was in charge of serine. These four are now purified and ready for cutting/inserting into our vector.
Daniel Steiner 17:08, 6 March 2014 (EST)
Today, we ran a gel of our PCR products from last time to make sure that we got the correct product.
Our lanes (from left to right): P, Q, R, S, T, ladder, ladder, V, W, Y
P has multiple bands, Q didn't show up at all, and R and T are both very faint, so we'll re-PCR these on Friday and try again.
Daniel Steiner 15:04, 4 March 2014 (EST)
Diluted our Thermus Thermophilus genomic DNA and the forward/reverse oligos for argS and serS. Set up PCR reactions using the "Basic PCR for Cloning" protocol. SerS and argS are both under 2kb, so both use the 2K55 PCR program.
Daniel Steiner 14:58, 27 February 2014 (EST)
Watched a demo of mini-prep and gel purification, then ran a mini-prep to purify our plasmid.
Daniel Steiner 13:41, 18 February 2014 (EST)
Designed forward/reverse oligos and made construction files for both argS and serS:
For argS, I added an NcoI site (incorporating the start codon) to the forward oligo, making sure to change the nucleotide after ATG to G, and an EcoR1 site to the reverse oligo.
- argS-F: cgttaCCATGGtacggcgcgcgctggaag
- argS-R: gcatcGAATTCttacattacctcaggcgcgg
For serS, I added BsmBI sites to both the forward and reverse oligos, so that they cut and create the same sticky ends that NcoI and EcoRI would create (CATG and AATT).
- serS-F: ctagaCGTCTCacatggtggacctcaagcgcctg
- serS-R: cactgCGTCTCgaattctatccgcacggctccagcac
Daniel Steiner 16:27, 14 February 2014 (EST)
Our group is in charge of finding the genes for the tRNA synthetases (M,N,P,Q,R,S,T,V,W,Y) in Thermus Thermophilus and creating oligos. We split up the amino acids among our group members, and I found the genes for arginyl-tRNA synthetase and seryl-tRNA synthetase.
ArgS has no internal NcoI or EcoRI restriction sites, but serS has an NcoI site (and BsaI) restriction site, so we'll have to use BsmBI to create the CATG sticky end or use soeing to introduce a silent mutation as we did in the tutorial.
We need to create the NcoI restriction site at the start of argS. It starts atgcta, when we need it to start atgG, so we can alter that single nucleotide (changing leucine to valine) when making the oligos.