SBB13Ntbk-LarryLi: Difference between revisions
No edit summary |
|||
| (4 intermediate revisions by the same user not shown) | |||
| Line 171: | Line 171: | ||
We restarted from digestion of PCA2 product. Followed same protocol as above. Purified digestion and put in freezer | We restarted from digestion of PCA2 product. Followed same protocol as above. Purified digestion and put in freezer | ||
==[[User:Larry Li|Larry Li]] | ==[[User:Larry Li|Larry Li]] 14:30, 4 April 2013 (EDT)== | ||
Redid ligation, transformation and plating. Note we did not add 50ul of water this time. | Redid ligation, transformation and plating. Note we did not add 50ul of water this time. | ||
==[[User:Larry Li|Larry Li]] 13:11, 9 April 2013 (EDT)== | |||
Picked colonies. Added 5 ml of LB + AMP then picked colonies and dropped into culture. Picked 4 colonies. | |||
Set to incubate overnight at 37C. | |||
==[[User:Larry Li|Larry Li]] 13:30, 18 April 2013 (EDT)== | |||
Purified DNA from cells using miniprep and then ran gel to check for band and presence of insert. | |||
Selected two best colonies for synthesis. | |||
==[[User:Larry Li|Larry Li]] 12:56, 16 April 2013 (EDT)== | |||
Analyzed sequencing data and selected correct vectors. | |||
==[[User:Larry Li|Larry Li]] 13:15, 23 April 2013 (EDT)== | |||
Transformed, rescued and plated cells.Followed previous transformation protocol but rescued for 1 hour. | |||
==[[User:Larry Li|Larry Li]] 13:20, 25 April 2013 (EDT)== | |||
Performed Miniprep, ran analytic gel and sent for sequencing. | |||
==[[User:Larry Li|Larry Li]] 13:04, 2 May 2013 (EDT)== | |||
Analyzed sequence data | |||
Latest revision as of 10:04, 2 May 2013
~~!~~
Larry Li 14:47, 5 March 2013 (EST)
CCOMT3 Construction File
Mix oligos (1-16) for one synthon, do PCA1 protocol (453 bp, pca1) PCR pca1 with external oligos (Newfoward and Newreverse) by PCA2 protocol (407 bp, pca2) Digest pca2 with EcoRI/BglII, gp (391+11+5 bp, pcr_dig) (get pre-digested vector from JCA of pBca9145-Bca1144) (EcoRI/BamHI, vect_dig) Ligate pcr_dig and vect_dig Oligo List: >CCOMT-3_oligo_1 TTTGGAATAGAAACAAACATGTCACCTCCGACATGTTGAGACGATGCAGATCTC >CCOMT-3_oligo_2 TTCCTTACCACCTGGATTGTGGGCTAACATGATTACGTCAATGTGGACTACAC >CCOMT-3_oligo_3 TTAGCCCACAATCCAGGTGGTAAGGAAAGAACACAGAAGGAGTTTGAAGACTT >CCOMT-3_oligo_4 GGCTTTGCCTGATAATGGTAAAGTTATTGTTGCCGAGTGCATTTTACCTGTCG >CCOMT-3_oligo_5 GGCCAAGGGTGCAGGTTTCCAGGGATTCAAGGTACATTGTAACGCATTTAATA >CCOMT-3_oligo_6 CTTTAGTTGCTAAAGATGAATCTGGTGCGACAGGTAAAATGCACTCGGCAACA >CCOMT-3_oligo_7 CTGTCATGATTGGTCTGACGAACATTGCTTGAAATTCTTAAAGAATTGTTATGA >CCOMT-3_oligo_8 CCTTCTTTAAGAACTCCATAATGTATGTATTAAATGCGTTACAATGTACCTTG >CCOMT-3_oligo_9 CATACATTATGGAGTTCTTAAAGAAGGTTGGATCCTGAGACCTGAGACGGCAT >CCOMT-3_oligo_10 CACCAGATTCATCTTTAGCAACTAAAGGTGTAGTCCACATTGACGTAATCATG >CCOMT-3_oligo_11 CAATGTTCGTCAGACCAATCATGACAGATCCATTTCATAAAGACGGCATCAGCC >CCOMT-3_oligo_12 ATATAGATGCCGTCCTAGCGAATTCATGAGATCTGCATCGTCTCAACATGTCGG >CCOMT-3_oligo_13 ATAACTTTACCATTATCAGGCAAAGCCTCATAACAATTCTTTAAGAATTTCAAG >CCOMT-3_oligo_14 AGGTGACATGTTTGTTTCTATTCCAAAGGCTGATGCCGTCTTTATGAAATGGAT >CCOMT-3_oligo_15 AATCCCTGGAAACCTGCACCCTTGGCCAAGTCTTCAAACTCCTTCTGTGTTCT >CCOMT-3_oligo_16 AAGTATCTTTCCTGTGCCCAGGATCCATGCCGTCTCAGGTCTCAGGATCCAA >CCOMT3-NewForward ccaaaGAATTCCGTCTCAACATGTCGGAGGTGAC >CCOMT3-NewReverse gactgAGATCTCGTCTCAGGTCTCAGGATCCAAC PCA1 Product: ATATAGATGCCGTCCTAGCGAATTCATGAGATCTGCATCGTCTCAACATGTCGGAGGTGACATGTTTGTTTCTATTCCAAAGGCTGATGCCGTCTTTATGAAATGGATCTGTCATGATTGGTCTGACGAACATTGCTTGAAATTCTTAAAGAATTGTTATGAGGCTTTGCCTGATAATGGTAAAGTTATTGTTGCCGAGTGCATTTTACCTGTCGCACCAGATTCATCTTTAGCAACTAAAGGTGTAGTCCACATTGACGTAATCATGTTAGCCCACAATCCAGGTGGTAAGGAAAGAACACAGAAGGAGTTTGAAGACTTGGCCAAGGGTGCAGGTTTCCAGGGATTCAAGGTACATTGTAACGCATTTAATACATACATTATGGAGTTCTTAAAGAAGGTTGGATCCTGAGACCTGAGACGGCATGGATCCTGGGCACAGGAAAGATACTT PCA2 Product: ccaaaGAATTCCGTCTCAACATGTCGGAGGTGACATGTTTGTTTCTATTCCAAAGGCTGATGCCGTCTTTATGAAATGGATCTGTCATGATTGGTCTGACGAACATTGCTTGAAATTCTTAAAGAATTGTTATGAGGCTTTGCCTGATAATGGTAAAGTTATTGTTGCCGAGTGCATTTTACCTGTCGCACCAGATTCATCTTTAGCAACTAAAGGTGTAGTCCACATTGACGTAATCATGTTAGCCCACAATCCAGGTGGTAAGGAAAGAACACAGAAGGAGTTTGAAGACTTGGCCAAGGGTGCAGGTTTCCAGGGATTCAAGGTACATTGTAACGCATTTAATACATACATTATGGAGTTCTTAAAGAAGGTTGGATCCTGAGACCTGAGACGAGATCTcagtc
On Thursday we will be doing the PCA.
Larry Li 13:06, 12 March 2013 (EDT)
On Thursday we performed the PCA1 protocol
38 uL ddH2O
5 ul 10x expand buffer
5 ul 2mM dNTPs
1 ul oligo mixture (100uM total, mixture of oligos after combination of 100uM stocks)
0.75 ul Expand polymerase
Program (can run JCA/PCA1)
2 min initial denature at 94oC
30 sec denature at 94oC
30 sec anneal at 55oC [This should be the overlap temp of your oligos - vary as needed]
30 sec extension at 72oC
repeat 2-4 30 times total
Today we performed a zymo cleanup on the assembly reaction and will perform amplification
For the zymo cleanup we
Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction.
Transfer into the Zymo column (small clear guys)
spin through, discard waste.
Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol)
spin through, discard waste.
Add 200 uL of Zymo Wash Buffer
spin through, discard waste.
spin for 90 seconds, full speed to dry.
elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction
and for the amplification
1 ul each outer oligo (10 uM)
1 ul purified pca product
.5 ul phusion
10 ul 5x phusion buffer
5 ul 2mM dNTPs
32.5 ul H2O
Program
2 min initial denature at 94oC
30 sec denature at 94oC
30 sec anneal at 60oC [This should be high, as your outer oligos now have a huge overlap with the correct product]
30 sec extension at 68oC
repeat 2-4 30 times total
Larry Li 12:42, 14 March 2013 (EDT)
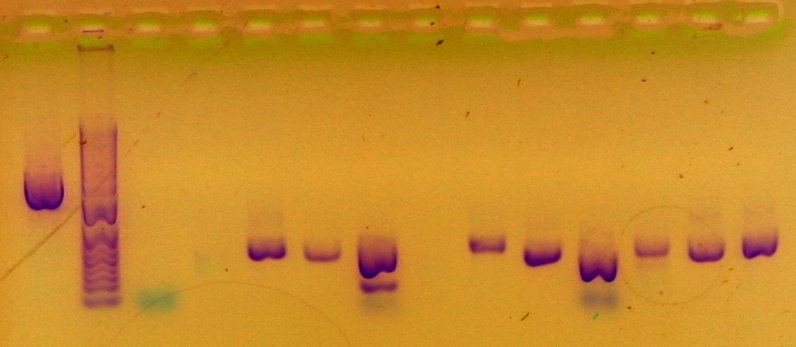
Sample was run in gel in lane 10.
Will perform zymo cleanup, digest and gel purify today.
Heated and dissolved gel in ADP buffer and put in freezer.
Larry Li 12:54, 19 March 2013 (EDT)
Finished zymo cleaning up today.
Larry Li 13:26, 21 March 2013 (EDT)
Today we performed ligation, transformation and plating.
We did
- Set up the following reaction:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase
- Pound upside down on the bench to mix
- Give it a quick spin to send it back to the bottom of the tube
- Incubate on the benchtop for 30min
- Put on ice and proceed to the transformation
Then
1. Thaw a 200 uL aliquot of cells on ice 2. Add 50 uL of water to the cells (if greater volume is desired) 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (for Miniprep product, dilute by 10, then use 1uL of dilution) 5. Add 70 uL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 90 seconds at 42 (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100uL of 2YT 10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight
Larry Li 12:31, 2 April 2013 (EDT)
Troubleshoot
We restarted from digestion of PCA2 product. Followed same protocol as above. Purified digestion and put in freezer
Larry Li 14:30, 4 April 2013 (EDT)
Redid ligation, transformation and plating. Note we did not add 50ul of water this time.
Larry Li 13:11, 9 April 2013 (EDT)
Picked colonies. Added 5 ml of LB + AMP then picked colonies and dropped into culture. Picked 4 colonies. Set to incubate overnight at 37C.
Larry Li 13:30, 18 April 2013 (EDT)
Purified DNA from cells using miniprep and then ran gel to check for band and presence of insert. Selected two best colonies for synthesis.
Larry Li 12:56, 16 April 2013 (EDT)
Analyzed sequencing data and selected correct vectors.
Larry Li 13:15, 23 April 2013 (EDT)
Transformed, rescued and plated cells.Followed previous transformation protocol but rescued for 1 hour.
Larry Li 13:20, 25 April 2013 (EDT)
Performed Miniprep, ran analytic gel and sent for sequencing.
Larry Li 13:04, 2 May 2013 (EDT)
Analyzed sequence data