SBB12Ntbk-Karen D. Alvarez
From OpenWetWare
~~!~~
~~!~~
Karen D. Alvarez 14:43, 16 February 2012 (EST)
==PCR for part sbb1217== 1. ca998/osbb1333R on template pBca9145-jtk2768 a)osbb1333R oligo is 34.6nM, MW=11,944.7, Tm=72.1°C. I prepared a dilute solution of oligos of 10μM. b) I mixed the following: 24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo ca998, 10uM 1uL Oligo osbb1333R, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA c)Labeled tube 2. osbb1333F/g00101 on template pBca9525-Bca1832 a)osbb1333F oligo is 34.0nM, MW=6,766.4, Tm=60.4°C. I prepared a dilute solution of 10μM. b) I mixed the following: 24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo g00101, 10uM 1uL Oligo osbb1333F, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA c)Labeled tube 3. Place oligos in ice bucket for PCR program according to predicted PCR product base pair length. a)ca998/osbb1333R PCR product is 643 bp b) osbb1333F/g00101 PCR product is 1438 bp ==PCA for sbb1206 part (leucine part)== 1. PCA1 on o1,o6,o2 (pca1) 2.I mixed 1μL of each oligo to a tube 3. I prepared the following mixture: 38 uL ddH2O 5 ul 10x expand buffer 5 ul 2mM dNTPs 1 ul oligo mixture 0.75 ul Expand polymerase 4. Labeled tube 5. Place in appropriate ice bucket for PCR
Karen D. Alvarez 23:54, 17 February 2012 (EST)
1. Obtained PCR product A and PCR product B. For each PCR product, loaded 6μL of PCR product with 4μL loading buffer in a single well of a 1% agarose gel. Run gel "gel purify" the products. 2. While gel was running, proceeded to do small-Frag zymo clean up(PCR reaction for fragments smaller than 300bp): a. Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction PCA1 b. Transfer into the Zymo column (small clear guys) c. Add 500uL of Ethanol and pipette up and down to mix d. spin through (15s), discard waste. e. Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) f. spin through (15s), discard waste. g. Add 200 uL of Zymo Wash Buffer h. spin through, discard waste. i. spin for 90 seconds, full speed to dry. j. elute with water (spin 60s) into a fresh Eppendorf tube 3. Continue with PCA amplification step using the purified PCA1 product. Prepared the following mixture to obtain PCA2 set up: 1 μL each outer oligo (10 uM): oligos o1 and o2 1 μL purified pca product: purified PCA1 from previous step 0.5 μL phusion 10 μL 5x phusion buffer 5 μL 2mM dNTPs 32.5 μL ddH2O 4. Perform PCR on PCA2 solution with the following program: a. 2 min initial denature at 94°C b. 30 sec denature at 94°C c. 30 sec anneal at 60°C [This should be high, as your outer oligos now have a huge overlap with the correct product] d. 30 sec extension at 68°C e. repeat 2-4 30 times total 5. After gel finished running bands were observed using UV light. Bands roughly correspond to their respective sizes (PCR product A=643 bp and B=1438 bp) ***Attached below*** a. Cut the bands and place in a tube b. Add about 650 μL of ADB buffer c. Place tube in water bath to melt the agarose d. Put away in freezer for further manipulations in NEXT lab class.
Karen D. Alvarez 14:45, 21 February 2012 (EST)
1. Continued with SOEing reaction of part ssb1217. I did Zymo Gel Purification procedure. a. Obtain Eppendorf tube containing the two gel fragments with buffer. b. Transfer into the Zymo column inside a collection tube (small clear guys) c. Spin 15 seconds, discard waste. d. Add 200 uL of Zymo Wash Bbuffer (which is basically 70% ethanol) e. Spin 15 seconds, discard waste. f. Add 200 uL of Zymo Wash Buffer g. spin through, discard waste. h. spin for 90 seconds, full speed to dry. i. Elute with 50 μL water (supposed to be 33 μL ) into a fresh Eppendorf tube 2. Set up the second PCR reaction for the purified eluded solution above: 24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo ca998, 10uM 1uL Oligo g00101, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA (eluded solution) a. Run PCR program as follows: 2 distinct cycling phases—the first 10 cycles have shorter extensions than the latter 20 cycles. You choose which one you want based on the length of your predicted product. If it is under 2kb, use 2K55. If it is between 2kb and 4kb, use 4K55. If it is over 4kb, use 8K55. In each case, start with the "55" version of the program. If you get no product or poor yield of product, repeat the same PCR reaction as two different variants, both with the “45” program. The first reaction will be the same composition as the first. For the second one, don’t add 3.3 uL of the water and instead use the volume for 3.3 uL of DMSO (dimethylsulfoxide) 3.Perform Small-Frag Zymo Cleanup on PCA2 product (leucine zipper part sbb1206 ~142 bp) (procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction for fragments smaller than 300bp. It also will remove the buffer and restriction enzymes from a restriction digest reaction) Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction Transfer into the Zymo column (small clear guys) Add 500uL of Ethanol and pipette up and down to mix spin through (15s), discard waste. Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) spin through (15s), discard waste. Add 200 uL of Zymo Wash Buffer spin through, discard waste. spin for 90 seconds, full speed to dry. elute with 50 μL water (spin 60s) into a fresh Eppendorf tube 4. Follow Digestion eluded PCA2 product NheI and BamHI as follows: 8μL of eluted PCA2 product 1μL of NEB Buffer 2 0.5 μL NheI 0.5 μL BamHI a. Incubate at 37°C on the thermocycler for 1hr b. Add this PCA2 to 100μL ADB buffer. c.Put away for NEXT lab
Karen D. Alvarez 15:00, 23 February 2012 (EST)
1. Obtain PCA2 digest product containing ADB buffer (from last lab class). Do small fragment zymo clean up as follows: a.Transfer into the Zymo column (small clear guys) b.Add 500uL of Ethanol and pipette up and down to mix c.spin through (15s), discard waste. d.Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) e.spin through (15s), discard waste. f.Add 200 uL of Zymo Wash Buffer g.spin through(15s), discard waste. h.spin for 90 seconds, full speed to dry. i.elute with 8μL ddwater (spin 60s) into a fresh Eppendorf tube 2. Do Regular Zymo Clean up on PCR product (A+B) as follows: a.Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction. b.Transfer into the Zymo column (small clear guys) c.spin through, discard waste. d.Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) e.spin through, discard waste. f.Add 200 uL of Zymo Wash Buffer g.spin through, discard waste. h.spin for 90 seconds, full speed to dry. e.elute with 33 μL water into a fresh Eppendorf tube 3.Digest "prcprd" on sbb1217 8uL of eluted PCR product 1uL of NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI a. Incubate at 37°C on the thermocycler for 1hr ( did 50 minutes) b. Run an agarose gel, and melt with 600μL ADB buffer at 55°C. 4. Perform Ligation reaction on PCA2 cleaned product (REDO this NEXT class)
Karen D. Alvarez 17:48, 24 February 2012 (EST)
1. Ligation reaction on PCA2 digest (1206dig + vectdig)as below: 6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase a. Pound upside down on the bench to mix b. Give it a quick spin to send it back to the bottom of the tube c. Incubate on the benchtop for 30min --> product is pBca9525-sbb1206 d. Put on ice and proceed to the transformation 2. Proceed to transformation by heat shock as follows: (Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock) 1. Thaw a 200 uL aliquot of cells on ice 2. Add 50 uL of water to the cells (if greater volume is desired) 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (for Miniprep product, dilute by 10, then use 1uL of dilution) 5. Add 70 uL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 90 seconds at 42 (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let shake in the 37°C incubator for 1 hour--did ~40min 10. Plate 75 uL on selective antibiotics, let incubate at 37°C overnight 3. Do zymo clean up on the gel purified pcrprd (A+B part sbb1217)
Karen D. Alvarez 15:18, 28 February 2012 (EST)
1. Since there was an inconsistent band in the SOEing pcrprd of part ss1217, redo this SOEing PCR reaction. a. Obtain the zymo cleaned A+B (after gel purification) and prepare the mixture below: 20.7uL ddH2O 3.3 uL DMSO 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo ca998, 10uM 1uL Oligo g00101, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA b. Run PCR reaction (pcrprd should be ~2056 bp long) 2. Redo Ligation reaction on leucine zipper PCA2 ss1206 6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest pBca9525-Bca1834 1uL Insert digest 0.5uL T4 DNA Ligase a. Pound upside down on the bench to mix b. Give it a quick spin to send it back to the bottom of the tube c. Incubate on the benchtop for 30min d. Put on ice and proceed to the transformation 3. Proceed to transform cells by heat-shock with ligation product -Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock. a. Thaw a 200 uL aliquot of cells on ice b. Add 30 uL of KCM to the cells c. Put your ligation mixture on ice, let cool a minute or two (for Miniprep product, dilute by 10, then use 1uL of dilution) d. Add 70 uL of the cell cocktail to the ligation, stir to mix e. Let sit on ice for 10 min f. Heat shock for 90 seconds at 42 (longer incubation may work better) g. Put back on ice for 1 min h. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour i. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight 4. Do mini-prep on the one cloudy tube, where bacteria grew. Procedure is as follows: a.Pellet 2 mL saturated culture by spinning full speed, 30 seconds in a 2mL Microcentrifuge tube. b.Dump supernatant c.Add 250uL of P1 buffer into each tube. Resuspend the cells thoroughly d.Add 250uL of P2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down. Solution should become clearer. e.Add 350uL of N3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then shake. f.Spin in centrifuge at top speed for 5 minutes. g.Label blue columns with an alcohol-resistant lab pen. h.Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds. i.Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin) j.Wash each column with 500 uL of PB buffer. k.Spin in centrifuge at full speed for 15 seconds, then flick out the liquid again. l.Wash with 750uL of PE buffer (washes the salts off the resins). m.Spin in centrifuge at full speed for 15 seconds and flick out liquid again. n.Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol. o.Label new Microcentrifuge tubes and put columns in them. p.Elute them by adding 50uL of water down the middle of the column (don't let it stick to the sides). q.Spin in centrifuge at top speed for 30 seconds. r.Take out columns and cap the tubes. *Clean up - note the P1 buffer is stored at 4degC and all the rest at room temperature.
Karen D. Alvarez 16:35, 1 March 2012 (EST)
1. Perform a regular zymo clean up on pcrpd_ssb1217 as follows: a.Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction. b.Transfer into the Zymo column (small clear guys) c.spin through 15 sec, discard waste. d.Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) e.spin through, discard waste. f.Add 200 uL of Zymo Wash Buffer g.spin through, discard waste. h.spin for 90 seconds, full speed to dry. i.Elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction (20.7 uL ddH2O 2.Digest pcrprd_sbb1217 a.Set up the following reaction: 8uL of eluted PCR product 1uL of NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI b.Incubate at 37°C on the thermocycler for 1hr c.Run an agarose gel(loaded 10uL of DNA +2 uL of dye) d.Cut band and melt with 600uL ADB buffer at 55°C. Note: Band observed looks the right size ~2059. Another band was observed of about 100 bp 3.Miniprep the samples obtained from transformation culture from last lab class. Note: 2 tubes: sbb1206-2 and sbb1206-3 a.Pellet 2 mL saturated culture by spinning full speed, 30 seconds in a 2mL Microcentrifuge tube. b.Dump supernatant c.Add 250uL of P1 buffer into each tube. Resuspend the cells thoroughly d.Add 250uL of P2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down. Solution should become clearer. e.Add 350uL of N3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then shake. f.Spin in centrifuge at top speed for 5 minutes. g.Label blue columns with an alcohol-resistant lab pen. h.Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds. i.Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin) j.Wash each column with 500 uL of PB buffer. k.Spin in centrifuge at full speed for 15 seconds, then flick out the liquid again. l.Wash with 750uL of PE buffer (washes the salts off the resins). m.Spin in centrifuge at full speed for 15 seconds and flick out liquid again. n.Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol. o.Label new Microcentrifuge tubes and put columns in them. p.Elute them by adding 50uL of water down the middle of the column (don't let it stick to the sides). q.Spin in centrifuge at top speed for 30 seconds. r.Take out columns and cap the tubes. 4. Keep 3 miniprep tubes( 1 from 02-28-12 and 2 from 03-01-12: sbb1206-1, sbb1206-2, and sbb1206-3)on the frigde.
Karen D. Alvarez 17:09, 2 March 2012 (EST)
1. Digest Miniprep tubes for the leucine zipper part sbb1206 (3 tubes). Prepare the following reaction:
2uL of miniprep
6uL of water
1uL of NEB Buffer 2
1uL BglII
a. Incubate at 37 degrees on the thermocycler for 1hr
b. Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an
acceptable stopping point*** If the DNA is shorter than 300bp, add 250uL of isopropanol and mix prior to loading it on the
column
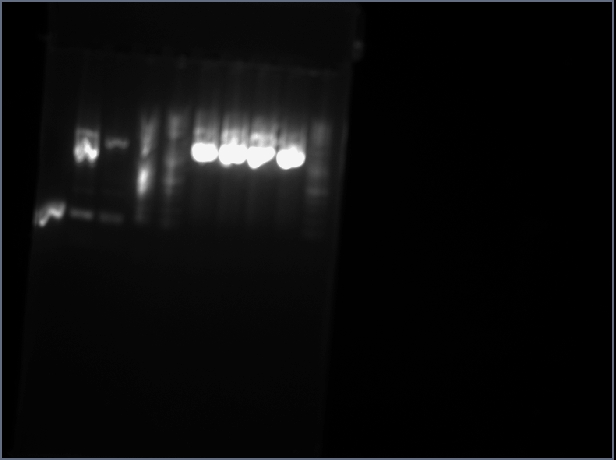
***From ApE prediction, digesting using this enzyme should give the following two bands: 2549 and 1071 bp
-The picture below shows the results of the analytical gel. Lanes 1 and 2 correspond to sbb1206 minipreps 1 and 2 respectively.These bands are consistent with the expected bands.
2. Do zymo clean up on digested pcrprd
a.transfer into the Zymo column inside a collection tube (small clear guys)
b.spin through 15 sec, discard waste.
c.Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol)
d.spin through, discard waste.
e.Add 200 uL of Zymo Wash Buffer
f.spin through, discard waste.
g.spin for 90 seconds, full speed to dry.
h.elute with water into a fresh Eppendorf tube-Label, this is ready for ligation reaction
Karen D. Alvarez 13:27, 6 March 2012 (EST)
1. Send part sbb1206 miniprep 1 and miniprep2 for sequencing. 2. Proceed to Ligation step of the digested part sbb1217. a.Set up the following reaction: 6.5uL ddH2O 1uL T4 DNA Ligase Buffer 1uL Vector digest (pBca9525-Bca1834) 1uL Insert digest 0.5uL T4 DNA Ligase b.Pound upside down on the bench to mix c.Give it a quick spin to send it back to the bottom of the tube d.Incubate on the benchtop for 30min e.Put on ice and proceed to the transformation 3. Transformation by heat-shock a. Thaw a 200 uL aliquot of cells on ice b. Add 50 uL of water to the cells (if greater volume is desired) c. Add 30 uL of KCM to the cells d. Put your ligation mixture on ice, let cool a minute or two (for Miniprep product, dilute by 10, then use 1uL of dilution) e. Add 70 uL of the cell cocktail to the ligation, stir to mix f. Let sit on ice for 10 min g. Heat shock for 10 min at 42 h. Put back on ice for 1 min i. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour j. Plate 75 uL on selective antibiotics, let incubate at 37 degrees overnight
Karen D. Alvarez 15:10, 8 March 2012 (EST)
1. Do the mini-prep on the transformed cells(part sbb1217) (4 tubes)
a.Pellet 2 mL saturated culture by spinning full speed, 30 seconds in a 2mL Microcentrifuge tube.
Dump supernatant
b.Add 250uL of P1 buffer into each tube. Resuspend the cells thoroughly
c.Add 250uL of P2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down.
Solution should become clearer.
d.Add 350uL of N3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to
precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then
shake.
e.Spin in centrifuge at top speed for 5 minutes.
f.Label blue columns with an alcohol-resistant lab pen.
g.Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds.
Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin)
h.Wash each column with 500 uL of PB buffer.
i.Spin in centrifuge at full speed for 15 seconds, then flick out the liquid again.
j.Wash with 750uL of PE buffer (washes the salts off the resins).
Spin in centrifuge at full speed for 15 seconds and flick out liquid again.
k.Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol.
l.Label new Microcentrifuge tubes and put columns in them.
Elute them by adding 50uL of water down the middle of the column (don't let it stick to the sides).
Spin in centrifuge at top speed for 30 seconds.
m.Take out columns and cap the tubes.
2. Prepare digestion mixture
1.5 uL of miniprep
6.5 uL of dH2O
1uL of NE Buffer 2
0.5uL EcoRI
0.5uL PstI
*This digestion enzymes should produce two bands on the gel of sizes 3030 and 1342 base pairs.
a.Incubate at 37 degrees on the thermocycler for 1hr
Karen D. Alvarez 13:51, 13 March 2012 (EDT)
1.Run gel of the 4 digested miniprep samples. Bands do not correspond to expected sizes. Gel is shown below:
Lanes:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Karen A sbb1217 mp1 | Karen A sbb1217 mp2 | Karen A sbb1217 mp3 | Karen A sbb1217 mp4 | Lader | sbb1224(1) | sbb1224(2) | AJ A+B | Karen A sbb1217 mp3 | Ladder |
Lanes:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| RO 1214 Dig | RO 1231 Dig | AJ A+B PCR digest | **** | Lader | KA | KA | KA | KA | ladder |
Karen D. Alvarez 14:36, 15 March 2012 (EDT)
1. Redo Digestion of miniprep using following volumes: 2. Run gel (shown below)
Lanes:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| sbb1217(1) | sbb1217(2) | sbb1217(3) | sbb1217(4) | ladder | ladder | -- | -- | -- | -- |