SBB12Ntbk-DanielChao: Difference between revisions
Daniel Chao (talk | contribs) |
Daniel Chao (talk | contribs) No edit summary |
||
| (17 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==[[User:Daniel Chao|Daniel]] 11:30, 16 February 2012 (PST)== | ==[[User:Daniel Chao|Daniel]] 11:30, 16 February 2012 (PST)== | ||
Prepared PCR reactions for sb1221: | Prepared PCR reactions for sb1221: | ||
| Line 16: | Line 14: | ||
Using protocol as described here: [[Template:SBB-Protocols_PCR1 | Wobble Reaction]] | Using protocol as described here: [[Template:SBB-Protocols_PCR1 | Wobble Reaction]] | ||
==[[User:Daniel Chao|Daniel]] 14:00, 17 February 2012 (PST)== | |||
Performed short-fragment Zymo on wobble product, using protocol as described here [[Template:SBB-Protocols_Zymo2 | Small-Frag Zymo Cleanup]]<br> | |||
==[[User:Daniel Chao|Daniel]] 12:30, 21 February 2012 (PST)== | |||
Ran gel of two PCR products used for SOEing [[SBB12_gels | Gel pics]] <br> Gel #1, lanes 3 and 4 pertain to fragments A (160bp) and B (1100bp) in the construction file for ssb1221. | |||
===Class Gel 1=== | |||
[[Image:2012_02_21_gel1_ssb2012spring.jpg]]<br> | |||
Lanes:<br> | |||
{| border="1" cellpadding="2" | |||
!width="1"|1 | |||
!width="1"|2 | |||
!width="1"|3 | |||
!width="1"|4 | |||
!width="1"|5 | |||
!width="1"|6 | |||
!width="1"|7 | |||
!width="1"|8 | |||
!width="1"|9 | |||
!width="1"|10 | |||
|- | |||
||Au PCA2||TN sbb1213||DC A||DC B||2-log ladder||JS||MM||JW||RO TP||RO CT | |||
|} | |||
Cut out gel and purified using [[Template:SBB-Protocols_Zymo3 | Zymo Gel Purification]]<br> | |||
Performed digest reaction on Zymo wobble product (1 hour in 37C), then purified using Small-Frag Zymo Cleanup. | |||
==[[User:Daniel Chao|Daniel]] 12:30, 23 February 2012 (PST)== | |||
Set up final PCR reaction for SOEing. | |||
Performed ligation reaction for sb-1228 Wobble products [[Template:SBB-Protocols_Enz4 | Ligation of EcoRI/BamHI digests]]<br> | |||
Heat-shock transformation into cells [[Template:SBB-Protocols_Micro1 | Transformation by heat-shock]]<br> | |||
Was not provided with enough cells (only about 100uL in tube). Added 30uL KCM to cells, and used 60uL of mixture for reaction. | |||
==[[User:Daniel Chao|Daniel]] 12:30, 24 February 2012 (PST)== | |||
Two things: | |||
1. Picked colonies from plate into four LB tubes for growth. <br> | |||
2. Ran gel on SOEing product to test for product presence, ran out of time to gel purify, so have to wait until next time. | |||
==[[User:Daniel Chao|Daniel]] 12:30, 28 February 2012 (PST)== | |||
Ran gel on SOEing product... not sure if products are present. | |||
===Class Gel #1=== | |||
[[Image:NEB_2-log_ladder.gif]][[Image:2012_03_1_gel1_ssb2012spring.jpg]]<br> | |||
The band is faint, and I'm not sure my SOEing product is reliable. I think it needs to be redone. | |||
No growth in four tubes -> JCA says no red colonies on plate is problem, so redo ligation reaction (may have picked wrong vector digest). So redid ligation reaction -> transformation -> plating. | |||
==[[User:Daniel Chao|Daniel]] 12:30, 1 March 2012 (PST)== | |||
Redid A+B SOEing reaction using CA998 + G00101 and A + B fragments. Wait until tomorrow to see results. | |||
Previously plated cells were picked (this time the plate had both red and white cells) and grew in LB tubes. Miniprep was performed on the resulting cell culture: [[Template:SBB-Protocols_Micro3 | Miniprep purification of DNA]]<br>. | |||
The purified DNA was stored. Analytical gel (mapping) will be run next time. | |||
==[[User:Daniel Chao|Daniel]] 12:30, 2 March 2012 (PST)== | |||
Previously miniprepped samples were digested using the protocol described here: [[Template:SBB-Protocols_Enz2 | EcoRI/BamHI Digest of PCR Products]] with BamHI and BglII enzymes. The protocol is a little off - used 1 uL of plasmid DNA (since high copy number). According to analysis using ApE, the resulting digest will contain a ~2400 bp band and a ~1600 bp band (if parent vector) or ~1000 bp band if desired product vector. This will enable us to differentiate between a parent vector w/ a mutation (explaining why the colony was white) and the part desired. The plasmids were digested, but not enough time for running the gel. | |||
The SOEing reaction was run on analytical gel and band was cut out for later Zymo gel purification. | |||
===Gel 1=== | |||
[[Image:NEB_2-log_ladder.gif]][[Image:2012_03_02_gel1_ssb2012spring.jpg]]<br> | |||
Lanes:<br> | |||
{| border="1" cellpadding="2" | |||
!width="1"|1 | |||
!width="1"|2 | |||
!width="1"|3 | |||
!width="1"|4 | |||
!width="1"|5 | |||
!width="1"|6 | |||
!width="1"|7 | |||
!width="1"|8 | |||
!width="1"|9 | |||
!width="1"|10 | |||
|- | |||
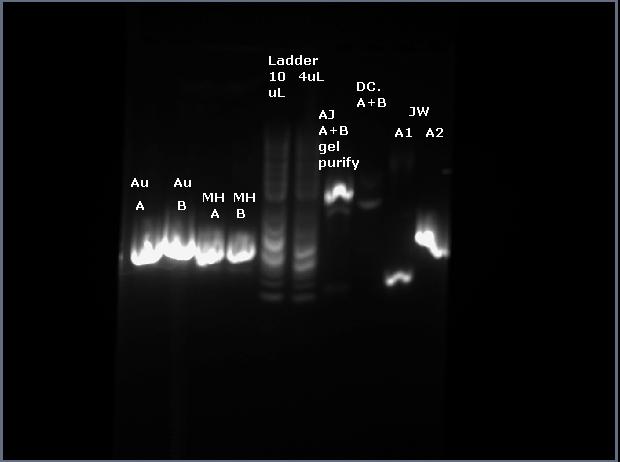
||Au SOE3||AJ A+B||empty||DC A+B||2log||empty||empty||empty||JW A2||2log | |||
|} | |||
The result from the gel was a little odd, however. It seems the ladder leaked throughout the gel, and could have contaminated my sample... I cut it (the heavier band) out nonetheless. | |||
==[[User:Daniel Chao|Daniel]] 12:30, 6 March 2012 (PST)== | |||
The BamHI BgII digest was run on analytical gel. Here are the results: | |||
===Gel 3=== | |||
[[Image:NEB_2-log_ladder.gif]][[Image:2012_03_05_gel3_ssb2012spring.jpg]]<br> | |||
It looks like the correct plasmid was obtained... the second band was about 1000 bp. | |||
I also performed [[Template:SBB-Protocols_Zymo3 | Zymo Gel Purification]] on the cut-out band from last week. I then ran a second gel to test whether I got what I was looking for. Interestingly, I got a funny result: | |||
===Class Gel 4=== | |||
[[Image:NEB_2-log_ladder.gif]][[Image:2012_03_06_gel4_ssb2012spring.jpg]]<br> | |||
Lanes:<br> | |||
{| border="1" cellpadding="2" | |||
!width="1"|1 | |||
!width="1"|2 | |||
!width="1"|3 | |||
!width="1"|4 | |||
!width="1"|5 | |||
!width="1"|6 | |||
!width="1"|7 | |||
!width="1"|8 | |||
!width="1"|9 | |||
!width="1"|10 | |||
|- | |||
||MM 1232-1||MM 1232-2||MM 1215-2||DC 1221 A+ B Pur||Ladder||JK 1233-2||JK 1204-1||JK 1204-2||PR 30-2||PR 30-3 | |||
|} | |||
==[[User:Daniel Chao|Daniel]] 12:30, 8 March 2012 (PST)== | |||
Entered both samples of sb1228 into Clotho and into plates for sequencing. | |||
Going to redo the SOEing PCR part from scratch. Produced four tubes - 2 with 33% water replaced by DMSO and 2 without, for each SOEing part. | |||
Latest revision as of 09:58, 13 March 2012
Daniel 11:30, 16 February 2012 (PST)
Prepared PCR reactions for sb1221:
PCR ca998/gfRevPR on pBjk2741-jtk2801 (160bp, A)
Placed in 100-500 PCR tray
PCR dc5002/g00101 on pBgl00001-Brp0006 (1100bp, B)
Placed in 1K-2K PCR tray
Using protocol as described here: Cloning by PCR
Prepared Wobble reaction for sbb1228
Wobble dc5003/dc5004 (64bp, wobpdt)
Using protocol as described here: Wobble Reaction
Daniel 14:00, 17 February 2012 (PST)
Performed short-fragment Zymo on wobble product, using protocol as described here Small-Frag Zymo Cleanup
Daniel 12:30, 21 February 2012 (PST)
Ran gel of two PCR products used for SOEing Gel pics
Gel #1, lanes 3 and 4 pertain to fragments A (160bp) and B (1100bp) in the construction file for ssb1221.
Class Gel 1
Lanes:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Au PCA2 | TN sbb1213 | DC A | DC B | 2-log ladder | JS | MM | JW | RO TP | RO CT |
Cut out gel and purified using Zymo Gel Purification
Performed digest reaction on Zymo wobble product (1 hour in 37C), then purified using Small-Frag Zymo Cleanup.
Daniel 12:30, 23 February 2012 (PST)
Set up final PCR reaction for SOEing.
Performed ligation reaction for sb-1228 Wobble products Ligation of EcoRI/BamHI digests
Heat-shock transformation into cells Transformation by heat-shock
Was not provided with enough cells (only about 100uL in tube). Added 30uL KCM to cells, and used 60uL of mixture for reaction.
Daniel 12:30, 24 February 2012 (PST)
Two things:
1. Picked colonies from plate into four LB tubes for growth.
2. Ran gel on SOEing product to test for product presence, ran out of time to gel purify, so have to wait until next time.
Daniel 12:30, 28 February 2012 (PST)
Ran gel on SOEing product... not sure if products are present.
Class Gel #1
The band is faint, and I'm not sure my SOEing product is reliable. I think it needs to be redone.
No growth in four tubes -> JCA says no red colonies on plate is problem, so redo ligation reaction (may have picked wrong vector digest). So redid ligation reaction -> transformation -> plating.
Daniel 12:30, 1 March 2012 (PST)
Redid A+B SOEing reaction using CA998 + G00101 and A + B fragments. Wait until tomorrow to see results.
Previously plated cells were picked (this time the plate had both red and white cells) and grew in LB tubes. Miniprep was performed on the resulting cell culture: Miniprep purification of DNA
.
The purified DNA was stored. Analytical gel (mapping) will be run next time.
Daniel 12:30, 2 March 2012 (PST)
Previously miniprepped samples were digested using the protocol described here: EcoRI/BamHI Digest of PCR Products with BamHI and BglII enzymes. The protocol is a little off - used 1 uL of plasmid DNA (since high copy number). According to analysis using ApE, the resulting digest will contain a ~2400 bp band and a ~1600 bp band (if parent vector) or ~1000 bp band if desired product vector. This will enable us to differentiate between a parent vector w/ a mutation (explaining why the colony was white) and the part desired. The plasmids were digested, but not enough time for running the gel.
The SOEing reaction was run on analytical gel and band was cut out for later Zymo gel purification.
Gel 1
Lanes:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Au SOE3 | AJ A+B | empty | DC A+B | 2log | empty | empty | empty | JW A2 | 2log |
The result from the gel was a little odd, however. It seems the ladder leaked throughout the gel, and could have contaminated my sample... I cut it (the heavier band) out nonetheless.
Daniel 12:30, 6 March 2012 (PST)
The BamHI BgII digest was run on analytical gel. Here are the results:
Gel 3
It looks like the correct plasmid was obtained... the second band was about 1000 bp.
I also performed Zymo Gel Purification on the cut-out band from last week. I then ran a second gel to test whether I got what I was looking for. Interestingly, I got a funny result:
Class Gel 4
Lanes:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| MM 1232-1 | MM 1232-2 | MM 1215-2 | DC 1221 A+ B Pur | Ladder | JK 1233-2 | JK 1204-1 | JK 1204-2 | PR 30-2 | PR 30-3 |
Daniel 12:30, 8 March 2012 (PST)
Entered both samples of sb1228 into Clotho and into plates for sequencing.
Going to redo the SOEing PCR part from scratch. Produced four tubes - 2 with 33% water replaced by DMSO and 2 without, for each SOEing part.