SBB12Ntbk-AuDuong: Difference between revisions
No edit summary |
No edit summary |
||
| Line 759: | Line 759: | ||
4) Do gel purification of digested SOE | 4) Do gel purification of digested SOE | ||
protocol used: | protocol used: | ||
1)cut out bands minimizing extra gel matter. | |||
2)put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle). | |||
3)heat in hand, shake until the gel has dissolved. | |||
4)do not add isopropanol because DNA is >300bp | |||
5)transfer into the Zymo column inside a collection tube (small clear guys) | |||
6)spin through, discard waste. | |||
7)Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) | |||
8)spin through, discard waste. | |||
9)Add 200 uL of Zymo Wash Buffer | |||
10)spin through, discard waste. | |||
11)spin for 90 seconds, full speed to dry. | |||
12)elute with water into a fresh Eppendorf tube | |||
Revision as of 14:48, 8 March 2012
~~!~~
*Au Duong 14:30, 16 February 2012 (EST)-THURSDAY:
1) Set up PCR and PCA reactions to create parts
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A)
***in practice, tube was labeled on top as "Au A" and on side as "AuD A 2/16/12"
PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B)
***in practice, tube was labeled on top as "Au B" and on side as "AuD B 2/16/12"
--> both PCR tubes were categorized as PCR 500bp-1kb
protocol used for setup:
resuspend oligos to make 100uM concentrations: - osbb1333R had 34.6nmoles; added 346uL ddH2O - osbb1229F had 29.4nmoles; added 294uL ddH2O dilute oligos to make 10uM concentrations (for use in this PCR protocol): - 9uL of ddH2O - 1uL of 100uM oligo Set up the following reaction in a PCR tube: 24uL ddH2O 3.3uL 10x Expand Buffer "2" NOTE: expand buffer is expand buffer, the "2" isn't very relevant 3.3uL dNTPs (2mM in each) 1uL Oligo 1, 10uM 1uL Oligo 2, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
PCA1 on o1,o8,o2 (pca1) ***in practice, tube was labeled on top as "Au pca1" and on side as "AuD PCA1 2/16/12"
protocol used for setup:
38 uL ddH2O
5 ul 10x expand buffer
5 ul 2mM dNTPs
1 ul oligo mixture (100uM total, mixture of oligos after combination of 100uM stocks)
(used .33 of each primer: o1,o2,o3; each at 100uM concentration)
0.75 ul Expand polymerase
*Au Duong 16:30, 17 February 2012 (EST)-FRIDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B)
1) Run PCR products on gel (10uL of PCR product +3uL of loading dye, 6uL of ladder) picture of gel (mine are the two left most wells)
2) Cut out DNA from gel and dissolve in ADB buffer (the brown zymo buffer bottle; to dissolve just warmed in hands and shook the the tube); not enough time to complete gel purification, so froze the dissolved gel/buffer solution
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
2) Clean up PCA1 reaction using zymo (small fragment variant of protocol) protocol used for setup:
1. Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction. 2. Transfer into the Zymo column (small clear guys) 3. Add 500uL of Ethanol and pipette up and down to mix 4. spin through (15s), discard waste. 5. Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) 6. spin through (15s), discard waste. 7. Add 200 uL of Zymo Wash Buffer 8. spin through, discard waste. 9. spin for 90 seconds, full speed to dry. 10. elute with 59uL water (spin 60s) into a fresh Eppendorf tube
3) Set up second PCA reaction PCA2 on o1,o2 (pca2) ***in practice, tube was labeled on top as "Au pca2" and on side as "AuD PCA2 2/17/12"
protocol used for setup:
32.5 uL ddH2O 10 ul 5x phusion buffer 5 ul 2mM dNTPs 1 ul oligo o1 (10mM) 1 ul oligo o2 (10mM) 1 ul template PCA1 (zymo purified) 0.5 ul Phusion polymerase
*Au Duong 14:33, 21 February 2012 (EST)-TUESDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B)
1) Do gel purification with gel dissolved in ADB buffer from last week
All spins until the drying step are 15 second full speed spins. 1)(did not add isopropanol b/c products are >300bp) 2) transfer into the Zymo column inside a collection tube (small clear guys) 3) spin through, discard waste. 4) Add 200 uL of Zymo Wash Bbuffer (which is basically 70% ethanol) 5) spin through, discard waste. 6) Add 200 uL of Zymo Wash Buffer 7) spin through, discard waste. 8) spin for 90 seconds, full speed to dry. 9) elute with 10uL water into a fresh Eppendorf tube (since I loaded 10uL into the gel originally) ***in practice, tube was labeled on top as "gel pur AuD A" and on side as "gel purified AuD PCR-A in 10uL H2O 2/21/12" and respectively for B
I also diluted the product 1:10 because for SOE-PCR less is more; too high of a concentration might oversaturate the PCR and give multiple bands
1uL of eluted PCR product 9uL of ddH2O ***in practice, tubes were labeled on top as "dil AuD A 1:10" and on side as "diluted AuD PCRA 2/21/12" and respectively for B
2) Set up SOE-PCR
PCR ca998/g00101 on A+B (1451bp, pcrpdt)
protocol used for setup:
24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo ca998, 10uM 1uL Oligo g00101, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA (A, diluted) 0.5uL Template DNA (B, diluted) ***in practice, tube was labeled on top as "Au SOE" and on side as "AuD SOE 2/21/12"
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
3) Run (unpurified)PCA2 reaction on on analytical gel

my pca2 is the left most sample
ladder used was 2log from NEB
I loaded 3uL of my PCA product +1.5uL of trypan blue (not the actual loading dye, TB is used for live/dead cell assays)
4) Clean up PCA2 reaction using zymo (small fragment variant of protocol) protocol used for setup:
1. Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction. 2. Transfer into the Zymo column (small clear guys) 3. Add 500uL of Ethanol and pipette up and down to mix 4. spin through (15s), discard waste. 5. Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) 6. spin through (15s), discard waste. 7. Add 200 uL of Zymo Wash Buffer 8. spin through, discard waste. 9. spin for 90 seconds, full speed to dry. 10. elute with 47uL water (spin 60s) into a fresh Eppendorf tube ***in practice, tube was labeled on top as "zymo clean AuD pca2" and on side as "zy clean AuD PCA2 in 47uL H2O 2/21/12"
5) Do Digestion with NheI/BamHI on PCA2(used PCR version of digestion, not wobble since from the gel it looked like I had a high concentration of product ) PCA2 on o1,o2 (pca2) ***in practice, tube was labeled on top as "Au dig pca2" and on side as "AuD dig PCA2 2/21/12"
Digest pca2 (NheI/BamHI, L, 1208dig) size is <142bp
protocol used for setup:
8uL of eluted PCR product 1uL of NEB Buffer 2 0.5uL NheI 0.5uL BamHI Incubate at 37*C in thermocycler for 1 hour
6) Stabilize digestion reaction with ADB buffer (not enough time to go through entire zymo purification protocol
Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction. ***in practice, 1.5mL tube was labeled on top as "Au dig pca2m in ADB" and on side as "AuD PCA2 dig in ADB 2/21/12"
*Au Duong 13:48, 23 February 2012 (EST)-THURSDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Do zymo cleanup (regular) with SOE PCR
All spins until the drying step are 15 second full speed spins. 1)(did not add isopropanol b/c products are >300bp) 2) transfer into the Zymo column inside a collection tube (small clear guys) 3) spin through, discard waste. 4) Add 200 uL of Zymo Wash Bbuffer (which is basically 70% ethanol) 5) spin through, discard waste. 6) Add 200 uL of Zymo Wash Buffer 7) spin through, discard waste. 8) spin for 90 seconds, full speed to dry. 9) elute with 33.6uL water into a fresh Eppendorf tube (since original PCR rxn was 33.6uL in volume) ***in practice, tube was labeled on top as "Au cleaned SOE" and on side as "zy clean SOE Au D in 33.6uL 2/23/2012"
2) Do digestion with EcoRI/BamHI on SOE-PCR (used slightly altered -for more product- PCR version of digestion, not wobble since from the gel it looked like I had a high concentration of product )
PCR ca998/g00101 on A+B (1451bp, pcrpdt) Digest pcrpdt (EcoRI/BamHI, L, pcrdig)
protocol used for setup:
16uL of eluted PCR product 2uL of NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI Incubate at 37*C in thermocycler for 1 hour ***in practice, tube was labeled on top as "Au dig SOE" and on side as "Au Dig SOE 2/23/12"
3) Run digestion product on gel (for later gel purification)
loaded 10uL of dig SOE + 3uL of loading dye
6uL of 2log ladder (NEB)
run at ~200V in small gel box
my sample is in the rightmost well
(even though the size looks suspicious, cut out bands and dissolve in 600uL of ADB buffer just in case
***in practice, tube was labeled on top as "dig SOE gel in ADB AuD" and on side as "AuD dig SOE gel in ADB 2/23/12"
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
4) Clean up digested PCA2(previously stablized in 100uL ADB buffer) using zymo (small fragment variant of protocol) protocol used for setup:
1. Transfer into the Zymo column (small clear guys) 2. Add 500uL of Ethanol and pipette up and down to mix 3. spin through (15s), discard waste. 4. Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) 5. spin through (15s), discard waste. 6. Add 200 uL of Zymo Wash Buffer 7. spin through, discard waste. 8. spin for 90 seconds, full speed to dry. 9. elute with 10uL water (spin 60s) into a fresh Eppendorf tube ***in practice, tube was labeled on top as "Au zy clean dig pca2" and on side as "AuD Nhe/Bam zymo clean dig PCA2 2/23/2012 in 10uL H2O"
5) Do ligation with digested PCA2 and vector plasmid
Digest pca2 (NheI/BamHI, L, 1208dig) Digest pBca9525-Bca1834 (NheI/BamHI, L, vectdig) Ligate 1208dig + vectdig, product is pBca9525-sbb1208
protocol used for setup:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase Incubate on the benchtop for 30min, then put on ice while waiting for competent cells ***in practice, tube was labeled on top as "AuD lig pca2" and on side as "AuD PCA2 lig 2/28/12"
6) Do transformation by heat shock with ligation mix and competent cells
1. Thaw a 200 uL aliquot of cells on ice 2. did not Add 50 uL of water to the cells (since each tube was only used by 1 student) 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (did not dilute since was ligation, not miniprep product) 5. Add 70 uL of the cell cocktail to the ligation, pipette stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 7 minutes 42*C (NOTE! the thermocycler was fitted with a .5mL size block, not PCR tube size, so there wasn't a perfect fit between tube and heating block) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let incubate in the 37 degree incubator for 1 hour 10. Plate 70+ uL on spec, let incubate at 37 degrees overnight
plate is labeled "spec AuD (PCA2) pBca9525-sbb1208 plated 2/23/2012"
*Au Duong 15:48, 24 February 2012 (EST)-FRIDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Redo SOE PCR twice(since according to the gel the product is of the correct size)
PCR ca998/g00101 on A+B (1451bp, pcrpdt)
protocol used for setup:
24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo ca998, 10uM 1uL Oligo g00101, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA (A, diluted) 0.5uL Template DNA (B, diluted) ***in practice, tube was labeled on top as "Au SOE1" and on side as "AuD SOE1 2/24/12" similarly for the 2nd tube
2) Discard the previous dissolved gel in ADB buffer (SOE dig) from yesterday; it's the wrong size
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
3) Since all the colonies look white, pick 4 large, isolated clonies from the spec plate "spec AuD (PCA2) pBca9525-sbb1208 plated 2/23/2012" streaked out yesterday --> plate had approx 20 large colonies and 10 small colonies
tubes are labeled with yellow tape on both caps and sides with "Au sbb1208 tube 1" and similarly for tubes 2-4 tubes have broth and spec antibiotic in them tubes will be chilled until Monday, where they'll be moved to an incubator to grow overnight
*Au Duong 12:57, 28 February 2012 (EST)-TUESDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Run SOE1 PCR on gel also run the original non diluted PCR-A and PCR-B to check the size loaded 10uL of SOE + 3uL of loading dye for SOE1 and SOE2 PCR loaded 1.8uL of PCR-A and PCR-B +3uL of loading dye 4uL of 2log ladder (NEB) run at ~190V in small gel box my samples are in the 4 leftmost wells (from L to R: A,B,1,2)
Discussion: even though the PCR-A and PCR-B were gel purified, there are still 2 bands showing up which might explain why the SOE failed again the larger of the two bands in A and B look to be the right size the brightest band in the SOE look to be ~2kb which is significantly larger than the 1.4kb of the expected product
2) Gel purify the first two bands (PCR-A and PCR-B)
cut out bands and dissolve in 600uL ADB buffer (not enough time to do entire gel purification procedure, just stabilize in ADB for now)
- in practice, tubes are labeled "Au D PCR A gel in ADB" on top and "Au D PCR A gel in ADB 2/28/12" on side of tube
similarly for B
3) Redo the first pcr reactions for soe pcr parts: Set up the following reaction in a PCR tube: PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A)
***in practice, tube was labeled on top as "Au A" and on side as "AuD A 2/28/12"
PCR osbbl229F/g00101 on pBca9525-bgl1031 (actually used 9525-Bgl1045 instead) (832bp, B)
***in practice, tube was labeled on top as "Au B" and on side as "AuD B 2/28/12"
24uL ddH2O 3.3uL 10x Expand Buffer "2" NOTE: expand buffer is expand buffer, the "2" isn't very relevant 3.3uL dNTPs (2mM in each) 1uL Oligo 1, 10uM 1uL Oligo 2, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
3) The RFP needs a while to show up, so it turns out that the larger colonies tend to be red (light red), and the smaller whites, so I picked all the wrong colonies last week it is also suspect because Prof. Anderson said I have a suspiciously small number of white colonies. Repick 4 colonies (white this time) and put into new spec+broth tubes
tubes are labeled with yellow tape on both caps and sides with "Au sbb1208 tube 1" and similarly for tubes 2-4 tubes have broth and spec antibiotic in them
4) Do ligation with digested PCA2 and vector plasmid
Digest pca2 (NheI/BamHI, L, 1208dig) Digest pBca9525-Bca1834 (NheI/BamHI, L, vectdig) Ligate 1208dig + vectdig, product is pBca9525-sbb1208
protocol used for setup:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase Incubate on the benchtop for 30min, then put on ice while waiting for competent cells
5) Do transformation by heat shock with ligation mix and competent cells
1. Thaw a 200 uL aliquot of cells on ice 2. did not Add 50 uL of water to the cells (since each tube was only used by 1 student) 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (did not dilute since was ligation, not miniprep product) 5. Add 70 uL of the cell cocktail to the ligation, pipette stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 2 minutes 42*C (NOTE! the thermocycler ) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let incubate in the 37 degree incubator for 1 hour 10. Plate 70+ uL on spec, let incubate at 37 degrees overnight
plate is labeled "spec AuD (PCA2) pBca9525-sbb1208 plated 2/28/2012"
*Au Duong 12:57, 1 March 2012 (EST)-THURSDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Run PCR-A and PCR-B (from yesterday)on gel for gel purification loaded 10uL of PCR-A and PCR-B + 3uL of loading dye (these are the PCR products I setup on 2/28/2012) 4uL and 10uL of 2log ladder (NEB) run at ~190V in small gel box my samples are in the 2 leftmost wells (from L to R: A,B)
Discussion: even though the PCR-A and PCR-B look to be the right size and single banded (although the band is very streaky)
2) Gel purify the first two bands (PCR-A and PCR-B)
cut out bands and dissolve in 600uL ADB buffer (not enough time to do entire gel purification procedure, just stabilize in ADB for now) ***in practice, tubes are labeled "Au D PCR A gel in ADB" on top and "Au D PCR A gel in ADB 3/1/12" on side of tube
similarly for B
3) Used the previously gel purified (done on 2/28/2012)PCR-A and PCR-B to do SOE-PCR protocol:
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A)
***in practice, tube was labeled on top as "Au A" and on side as "AuD A 2/28/12"
PCR osbbl229F/g00101 on pBca9525-bgl1031 (actually used 9525-Bgl1045 instead) (832bp, B)
***in practice, tube was labeled on top as "Au B" and on side as "AuD B 2/28/12"
24uL ddH2O 3.3uL 10x Expand Buffer "2" NOTE: expand buffer is expand buffer, the "2" isn't very relevant 3.3uL dNTPs (2mM in each) 1uL Oligo 1 (ca998) 10uM 1uL Oligo 2 (g00101) 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA (A and B)
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
4) Zach picked 3 more colonies from my 2 plates; of those 1 grew. Out of the 7 tubes (4 from me, 3 from Zach) 3 had significant growth (the other ones looked either dead or very slow growing); Do Mini prep (plasmid extraction) from the 3 that worked
protocol:
- Pellet 2 mL saturated culture by spinning full speed, 30 seconds in a 2mL Microcentrifuge tube.
- Dump supernatant
- Add 250uL of P1 buffer into each tube. Resuspend the cells thoroughly
- Add 250uL of P2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down. Solution should become clearer.
- Add 350uL of N3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then shake.
- Spin in centrifuge at top speed for 5 minutes.
- Label blue columns with an alcohol-resistant lab pen.
- Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds.
- Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin)
- Wash each column with 500 uL of PB buffer.
- Spin in centrifuge at full speed for 15 seconds, then flick out the liquid again.
- Wash with 750uL of PE buffer (washes the salts off the resins).
- Spin in centrifuge at full speed for 15 seconds and flick out liquid again.
- Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol.
- Label new Microcentrifuge tubes and put columns in them.
- Elute them by adding 50uL of water down the middle of the column (don't let it stick to the sides).
- Spin in centrifuge at top speed for 30 seconds.
- Take out columns and cap the tubes.
***in practice tubes are labeled "Au D Ec1 sbb1208 leu" on top and "AuD Ec1 sbb1208 leu 3/1/2012" on the side; simiarly for strains Ec2 and Ec3
*Au Duong 15:59, 2 March 2012 (EST)-FRIDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Run SOE3 PCR on gel also run the original non diluted PCR-A and PCR-B to check the size
loaded 10uL of SOE + 3uL of loading dye for SOE3
4uL of 2log ladder (NEB)
run at ~190V in small gel box
my sample is in the leftmost well
Discussion: It looks like the SOE-PCR failed again (the band looks >>3kb when my expected product is only ~1.4kb)
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
2) Digest plasmids extracted from E coli yesterday (Ec1, Ec2, Ec3) and the parent plasmid pBca9525-Bca1834
Analysis: Digestion of parent vector pBca9525-Bca1834 with NheI/BamHI will yield: 3494+841 Digestion of good plasmid with NheI/BamHI will yield: 3494+142 (actually a bit less) Conclusion: The difference between parent and 'good' plasmids should be readily distinguishable on a gel when the plasmids are digested by NheI/BamHI
Protocol for Digestion : Note: this is adjusted for high copy number plasmids such as pBca9525
For Ec1, Ec2, Ec3 6uL ddH2O 2uL Miniprepped plasmid 1uL 10x NEB Buffer 2 .5uL NheI .5uL BamHI
For pBca9525-Bca1834 (aka "K" in the tube, for 'control') 7uL ddH2O 1uL Miniprepped plasmid 1uL 10x NEB Buffer 2 .5uL NheI .5uL BamHI
- Incubate at 37 on the thermocycler for 60 minutes
- In practice, tubes were labeled "Ec1" on top and "3/2/2012" on side, simiarlly for Ec2, Ec3 and K (K=control=parent plasmid)
NOTE: There was not enough time and gels to run the digested plasmids on a gel on the same day. Since these digestions will just be used for mapping (we send in the entire miniprepped plasmids for sequencing), freezing these samples and running them on a gel Monday should be okay (confirmed with Zach)
*Au Duong 14:05, 6 March 2012 (EST)-TUESDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Do Zymo cleanup (regular, not small fragment) on SOE3
PCR ca998/g00101 on A+B (1451bp, pcrpdt)
protocol used for setup:
- Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of Zymo Wash Buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction
***in practice, tube was labeled on top as "zymo clean AuD SOE3" and on side as "zy clean Au SOE3 in 40uL H2O 3/5/12"
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
2) Run Digested plasmids on gel along with parent vector digest control
Analysis: Digestion of parent vector pBca9525-Bca1834 with NheI/BamHI will yield: 3494+841 Digestion of good plasmid with NheI/BamHI will yield: 3494+142 (actually a bit less) Conclusion: The difference between parent and 'good' plasmids should be readily distinguishable on a gel when the plasmids are digested by NheI/BamHI
3) Rerun PCA2 product (from 2/21/2012, has been zymo cleaned)to check size (since the previous gel had a ugly ladder)
Discussion: the size looks a little bit bigger than 100bp, but I'm expecting (~140bp). Zach says the band might be 140bp anyways
*Au Duong 13:08, 8 March 2012 (EST)-THURSDAY:
For part sbb1229
Featurename: cI-TraR-B Genename: cI, traR Source: Lambda phage / Agrobacterium
PCR ca998/osbbl333R on pBca9145-jtk2768 (643bp, A) PCR osbbl229F/g00101 on pBca9525-bgl1031 (832bp, B) PCR ca998/g00101 on A+B (1451bp, pcrpdt)
1) Do Digestion with EcoRI/BamHI on SOE3 (used slightly altered -for more product- PCR version of digestion, not wobble since from the gel it looked like I had a high concentration of product )
PCR ca998/g00101 on A+B (1451bp, pcrpdt) Digest pcrpdt (EcoRI/BamHI, L, pcrdig)
protocol used for setup:
16uL of eluted PCR product 2uL of NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI Incubate at 37*C in thermocycler for 1 hour ***in practice, tube was labeled on top as "Au dig SOE" and on side as "Au Dig SOE 3/8/12"
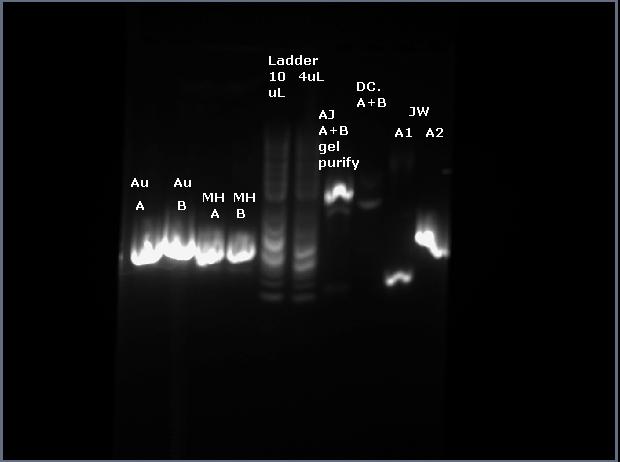
3) Run digestion product on gel (for later gel purification)
loaded 10uL of dig SOE + 3uL of loading dye
3uL of 2log ladder (NEB)
run at 200V in small gel box
my sample is in the rightmost well
4) Do gel purification of digested SOE
protocol used: 1)cut out bands minimizing extra gel matter. 2)put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle). 3)heat in hand, shake until the gel has dissolved. 4)do not add isopropanol because DNA is >300bp 5)transfer into the Zymo column inside a collection tube (small clear guys) 6)spin through, discard waste. 7)Add 200 uL of Zymo Wash Buffer (which is basically 70% ethanol) 8)spin through, discard waste. 9)Add 200 uL of Zymo Wash Buffer 10)spin through, discard waste. 11)spin for 90 seconds, full speed to dry. 12)elute with water into a fresh Eppendorf tube
***in practice, tube was labeled on top as "dig SOE gel in ADB AuD" and on side as "AuD dig SOE gel in ADB 2/23/12"
For part: sbb1208
Featurename: lz_EVLR Genename: leucine zipper variant Source: Synthetic, see PMID:12459719
5) Do ligation with digested PCA2 and vector plasmid
Digest pca2 (NheI/BamHI, L, 1208dig) Digest pBca9525-Bca1834 (NheI/BamHI, L, vectdig) Ligate 1208dig + vectdig, product is pBca9525-sbb1208
protocol used for setup:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase Incubate on the benchtop for 30min, then put on ice while waiting for competent cells ***in practice, tube was labeled on top as "lig Au pca2" and on side as "lig Au PCA2 3/8/12"
6) Do transformation by heat shock with ligation mix and competent cells
1. Thaw a 200 uL aliquot of cells on ice 2. did not Add 50 uL of water to the cells (since each tube was only used by 1 student) 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two (did not dilute since was ligation, not miniprep product) 5. Add 70 uL of the cell cocktail to the ligation, pipette stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 15 minutes 42*C (NOTE! the thermocycler was fitted with a .5mL size block, not PCR tube size, so there wasn't a perfect fit between tube and heating block) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let incubate in the 37 degree incubator for 1 hour 10. Plate 70+ uL on spec, let incubate at 37 degrees overnight
plate is labeled " AuD sbb1208 spec plated 3/8/2012"