SBB12Ntbk-Annie Joseph
Annie Joseph 16:23, 13 February 2012 (EST)
We did initial PCR/PCA today!
For part sbb1216:
Obtained and made a 10 micro molar solution of oligos ca998/osbb1333R and osbb1216F/g00101 for PCR.
Set up two solutions for PCR:
This one I call A in construction file:
24uL ddH2O
3.3uL 10x Expand Buffer "2"
3.3uL dNTPs (2mM in each)
1uL ca998, 10uM
1uL osbb1333R, 10uM
0.5uL Expand polymerase "1"
0.5uL pBca9145-jtk2768
And...
This one is B:
24uL ddH2O
3.3uL 10x Expand Buffer "2"
3.3uL dNTPs (2mM in each)
1uL osbb1216F, 10uM
1uL g00101, 10uM
0.5uL Expand polymerase "1"
0.5uL pBjk2741-jtk3346
For part sbb1205
I set up a solution based on the PCA protocol on open wet ware.
38 uL ddH2O
5 ul 10x expand buffer
5 ul 2mM dNTPs
1 ul oligo mixture (100uM total, mixture of oligos after combination of 100uM stocks)
0.75 ul Expand polymerase
Notes:
n/a
Annie Joseph 12:30, 17 February 2012 (EST)
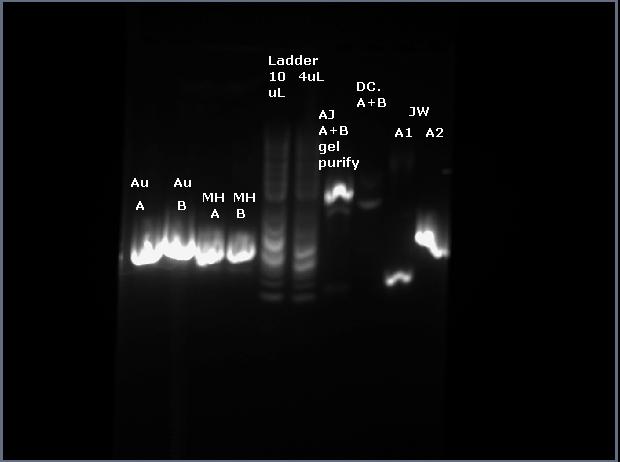
Ran two lanes on agarose gel for gel purification of products A and B from last time. Extracted the product A from the gel. The lane
for B was very faint so I have decided to redo the gel one more time before considering whether it was a mistake in an earlier step :(
Created the PCA solution for creation of pca2.
For next time: I will rerun the gels (PCR A&B with loading buffer), extract Create the digests for the part and template and use thermocycler (time for ligation ?)
Possibly with time remaining, I will set up the pcr reaction with A+B
Annie Joseph 12:30, 21 February 2012 (EST)
For the sbb1216 part, I ran the two PCR products from last week on the agarose gel for gel purification:
PCR ca998/osbb1333R on pBca9145-jtk2768 (A)
PCR osbb1216F/g00101 on pBjk2741-jtk3346 (B)
Obtained alright results, can be seen on the gel two lanes 8 and 9. The bands (brightest ones) look to be about the right size.
Cut out the bands and did the rest of the gel purification for both A and B.
For the Leucine Zipper part (sbb1205):
I had the pca2 product from last week, which I ran on gel for analysis (lane 4 ?) looks alright.
Did a cleanup of the pca2 product (did the protocol for regular zymo cleanup which I think might have been better to use the small frag
cleanup given the size of the pca2) and proceeded to do the digest of pca2 with NheI/BamHI following the protocol for EcoRI/BamHI
Digest of Wobble Products for the size considerations. Kept the digest on 37 degrees on the thermocycler for ~1 hour. Finally did a
zymo small fragment cleanup this time.
Annie Joseph 12:30, 23 February 2012 (EST)
sick :(
Annie Joseph 12:30, 24 February 2012 (EST)
Set up the A+B PCR with parts ca998/g00101 for my sbb1216 part
Same PCR protocol as before but there are two templates A and B from last tuesday that is supposed to anneal at their complementary
region as part of the Soeing PCR process
Also attempted a transformation but underestimated the time requirement
Ligated the PCA2 digest with NheI/BamHI with the Bca1834 vector digest NheI/BamHI.
Allowed the Ligate product to incubate for 30 minutes on benchtop.
Went through the protocol for transformation with 200 micro liters of cells.
- Last step modification: (not sure how it will affect the cells ability to have grown) only incubated the cells with ligate for around
35 minutes versus 1 hour after heat shock.
Annie Joseph 12:30, 28 February 2012 (EST)
Zach picked the colonies over the weekend and had them growing in tubes when we arrived today. He picked 4 colonies (white, the red
colonies indicates failure of transformation and there were some red colonies). Only one of my colonies out of 4 grew. I did miniprep
procedure to isolate the plasmid DNA from this colony of cells using the protocol listed in open wet ware. The extracted DNA has
been saved in a tube (eluted with water to a total volume of 50 micro liter). Now I need to digest and run an analytical gel before
deciding whether or not to sequence.
I also picked three more colonies from the plate to grow overnight to see if I have any success growing more, see if more of them
transformed.
For the other part I am working on (sbb1216) I got back the pcr product for the A+B pcr reaction with end oligos. Ran it on gel,
results will determine whether I can digest, ligate, etc.
Annie Joseph 12:30, 1 March 2012 (EST)
None of the picked colonies grew :(
So I have only one colony to work with which was miniprepped last time. Today I am running a digest of it to do an analytical gel analysis.
The analysis shows no bands on the gel lane (lane 7):
This is strange because there was no DNA present, which I think indicates a failure in the miniprep step. However, the transformation step was rushed, so I will go back and do another ligation/transformation.
Also have the A+B on gel to gel purify and set up a digest of A+B. The A+B band is too big (~3000 bp when it should be ~1500):
Annie Joseph 12:30, 6 March 2012 (EST)
The cell cultures picked from the last plating grew(4 colonies).
Did miniprep purification of the DNA from the 4 cell cultures. Hopefully there are no mistakes in this step and I have good plasmid DNA
in the 50 microliter eluted with water. One possible area of mistake is that I forgot to discard the PB buffer at the bottom of the
tube before adding PE and centrifuging so there was some PE remaining in the blue column which I centrifuged one more time after
discarding the flow.
The DNA obtained from the miniprep step was digested with NheI and BamHI using the protocol for analytical digest and incubated for
approximately 25-30 minutes. And ran on gel (Gel 5 lane 8, 9, 10). Did not run the digest from colony 4 (no space on gel).
I also ran the A+B soeing PCR product on gel and cut out the band (see Gel 2 lane 1).
Annie Joseph 12:30, 8 March 2012 (EST)
Did a zymo cleanup of the A+B gel purified product and eluted with 50 microliter of water. The cleaned up product A+B was digested with
EcoRI and BamHi and incubated on the thermocycler for one hour. Then it is run on the gel for cutting it out. Will cut out if there is
enough time, otherwise will have to repeat the digestion and gel running step the next time. I can still check to see whether the
bands are the right size.
Also redid the analytical gel for the 4 miniprep products from the isolated colonies from last time. The last gel(analytical) did not
look correct and I wanted to confirm whether it was because I used too much of the miniprepped product (used 4 microliters before while
I only used 1 microliter today). If the bands turn out okay,
sequencing. Otherwise, I will try picking other colonies and see if they grow, then miniprep if there is time.
Annie Joseph 12:30, 13 March 2012 (EST)
Last time, the gel for the A+B digest turned up nothing. I am reasoning that the A+B pcr product band I cut out from the gel (gel
purify) was too faint or the cleanup after extraction of the gel was enough to dilute it too much to not have anything show up when I
ran the digest and ran it through the gel again. (see lane 8, nothing!)
I am also running out of A+B pcr product, so I decided to save what I have (need to run it on gel and cut out) and run soeing PCR one
last time on the A and B pcrpdt to create A+B pcr product again.
The miniprep product's (for the Leucine Zipper parts) analytical gel turned out alright however. I was expecting a 3.5 kb template and
a 126 bp part bands on the gel when I cut with NheI/BamHI. Colonies 1,3,4 look approximately to be the correct band sizes on the
gel(see above gel lane 1,2,3,4 corresponding to respective colonies). The 3.5 kb band is rather thick and could be much larger but this
might again be an effect of the amount of DNA I loaded for the analytical gel (only used 1 micro liter b/c of the high copy number of
the plasmid) I gave these to be sequenced (only supposed to give 2 max, accidentally gave 3 now of the same part but different
colonies!) and input into clotho as part sbb1205.
Annie Joseph 12:30, 15 March 2012 (EST)
Got the third A+B soeing PCR product. Ran on gel. Size seems to be about right. Cut out the band and kept it in ADB buffer for next
time.