SBB10Ntbk-XiaoY.Liu: Difference between revisions
Xiao Y. Liu (talk | contribs) |
Xiao Y. Liu (talk | contribs) No edit summary |
||
| (44 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 17:40, 21 April 2010 (EDT)== | |||
* Team 1 project | |||
* Link to team notebook [[SBB10AssayTeam1-LabNotebook]] | |||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:06, 19 March 2010 (EDT)== | |||
===PiggyBac=== | |||
* miniprep and mapping | |||
* stop at incubation at 37°C, Mike will run the gel. | |||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:09, 17 March 2010 (EDT)== | |||
===PiggyBac=== | |||
* Gel Image of digested product | |||
[[Image:WCD 20100310.jpg]] | |||
*zymo gel purification of digested product. elute with 8ul water. | |||
* Ligation and Transformation. | |||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:10, 10 March 2010 (EST)== | |||
===PiggyBac=== | |||
* obtained SOEing PCR product back, do an analytical gel(3ul pcr, 7ul loading buffer) | |||
[[Image:xiao's piggybac SOEing porduct.jpg]] | |||
* zymo clean up. elute with 30ul water | |||
====Regular Zymo Cleanup==== | |||
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction. | |||
#Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction. | |||
#Transfer into the Zymo column (small clear guys) | |||
#spin through, discard waste. | |||
#Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol) | |||
#spin through, discard waste. | |||
#Add 200 uL of PE or Zymo Wash buffer | |||
#spin through, discard waste. | |||
#spin for 90 seconds, full speed to dry. | |||
#elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction | |||
* digestion | |||
====EcoRI/BamHI Digest of PCR Products==== | |||
'''For PCR products, you will only digest a portion of your purified PCR product. Note that you must make a minor modification to the procedure if your DNA is shorter than 300bp (you add a little isopropanol to the ADB after melting).''' | |||
*Set up the following reaction: | |||
8uL of eluted PCR product | |||
1uL of NEB Buffer 2 | |||
0.5uL EcoRI | |||
0.5uL BamHI | |||
*Incubate at 37 degrees on the thermocycler for 1hr | |||
*Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an acceptable stopping point | |||
*If the DNA is shorter than 300bp, add 250uL of isopropanol and mix prior to loading it on the column | |||
* dut to time manner, ask Will to do the gel and buffer for me. | |||
=== sbb03-tos=== | |||
* ligation and transformation | |||
* obtained digested product from professor. | |||
====Ligation of EcoRI/BamHI digests==== | |||
*Set up the following reaction: | |||
6.5uL ddH2O | |||
1uL T4 DNA Ligase Buffer (small red or black-striped tubes) | |||
1uL Vector digest | |||
1uL Insert digest | |||
0.5uL T4 DNA Ligase | |||
*Pound upside down on the bench to mix | |||
*Give it a quick spin to send it back to the bottom of the tube | |||
*Incubate on the benchtop for 30min | |||
*Put on ice and proceed to the transformation | |||
====Transformation by heat-shock==== | |||
'''Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock.''' | |||
1. Thaw a 200 uL aliquot of cells on ice | |||
2. Add 50 uL of water to the cells | |||
3. Add 30 uL of KCM to the cells | |||
4. Put your ligation mixture on ice, let cool a minute or two | |||
5. Add 70 uL of the cell cocktail to the ligation, stir to mix | |||
6. Let sit on ice for 10 min | |||
7. Heat shock for 90 seconds at 42 (longer incubation may work better) | |||
8. Put back on ice for 1 min | |||
9. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour | |||
10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight | |||
==[[User:JCAnderson|JCAnderson]] 18:59, 9 March 2010 (EST)== | |||
[[Image:JCA031010gel1.jpg|150px|right]] | |||
JCA reran the PCA using phusion instead of expand in step 1, came out much better (gel at right). Took through to digestion and cleanup, ready to ligate. | |||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:16, 8 March 2010 (EST)== | ==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:16, 8 March 2010 (EST)== | ||
Xiao's gel on March 8th | Xiao's gel on March 8th | ||
| Line 46: | Line 125: | ||
====Amplification==== | ====Amplification==== | ||
Now, you need to do an amplification of the correct full-length chunks. Clean up the assembly reaction with a zymo column; don't bother running it on a gel - it'll be a smeary mess and won't really help you. | Now, you need to do an amplification of the correct full-length chunks. Clean up the assembly reaction with a zymo column; don't bother running it on a gel - it'll be a smeary mess and won't really help you. | ||
====Regular Zymo Cleanup==== | |||
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction. | The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction. | ||
| Line 99: | Line 178: | ||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:21, 24 February 2010 (EST)== | ==[[User:Xiao Y. Liu|Xiao Y. Liu]] 16:21, 24 February 2010 (EST)== | ||
===PiggyBac=== | ===PiggyBac=== | ||
==== Second PCR trial for PiggyBac A ==== | |||
* PCR for piggyBac A failed, so do a second PCR with oligos(ca1530 and piggyBac02) and with program 55. | * PCR for piggyBac A failed, so do a second PCR with oligos(ca1530 and piggyBac02) and with program 55. | ||
make dilution(10uM) puggyBac01 from stock(100uM) with 9ul water and 1ul stock | make dilution(10uM) puggyBac01 from stock(100uM) with 9ul water and 1ul stock | ||
| Line 114: | Line 193: | ||
0.5uL Expand polymerase "1" | 0.5uL Expand polymerase "1" | ||
* place them in 55 PCR program. | * place them in 55 PCR program. | ||
==== | ==== PiggyBac B and PiggyBac C==== | ||
* after preparative gel, zymo purification. | * after preparative gel, zymo purification. | ||
=====Zymo Gel Purification===== | =====Zymo Gel Purification===== | ||
*All spins until the drying step are 15 second full speed spins. | *All spins until the drying step are 15 second full speed spins. | ||
| Line 133: | Line 213: | ||
* contiune sbb03 experiment. PCR with tos-1 and tos-20 | * contiune sbb03 experiment. PCR with tos-1 and tos-20 | ||
* add following in order: (check order with Prof. ) | * add following in order: (check order with Prof. ) | ||
32.5 ul H2O | |||
10 ul 5x phusion buffer | |||
5 ul 2mM dNTPs | |||
1 ul each outer oligo (10 uM)=tos-1 and tos-20 | |||
0.5 ul phusion | |||
* run Phusion Program, anneal at 60°C, | * run Phusion Program, anneal at 60°C, | ||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 19:07, 22 February 2010 (EST)== | ==[[User:Xiao Y. Liu|Xiao Y. Liu]] 19:07, 22 February 2010 (EST)== | ||
===PiggyBac=== | ===PiggyBac=== | ||
* Products from 3 PCR for piggyBac, do a preparative gel with sbb04A, sbb04B, and sbb04C with 6ul PCR products and 4ul loading buffer(blue, dye). | |||
* sbb04 stops at melting gel with ADP buffer. | * sbb04 stops at melting gel with ADP buffer. | ||
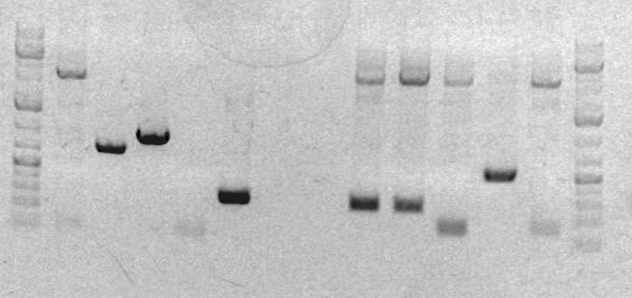
Image from preparative gel | Image from preparative gel | ||
| Line 153: | Line 231: | ||
1. ladder 2. sbb04A 3. sbb04B 4. sbb04C 5. RH 33A 6. RH33B 7. blank 8. blank 9. CD1 10. CD2 11. CD3 12. sbb19A 13. sbb19B 14. ladder | 1. ladder 2. sbb04A 3. sbb04B 4. sbb04C 5. RH 33A 6. RH33B 7. blank 8. blank 9. CD1 10. CD2 11. CD3 12. sbb19A 13. sbb19B 14. ladder | ||
== | === sbb03 tos=== | ||
* After getting PCA products back for sbb03, do a zymo clean up | * After getting PCA products back for sbb03, do a zymo clean up. | ||
====Regular Zymo Cleanup==== | |||
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction. | |||
#Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction. | |||
#Transfer into the Zymo column (small clear guys) | |||
#spin through, discard waste. | |||
#Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol) | |||
#spin through, discard waste. | |||
#Add 200 uL of PE or Zymo Wash buffer | |||
#spin through, discard waste. | |||
#spin for 90 seconds, full speed to dry. | |||
#elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction (500ul) | |||
* sbb03 stops at the end of zymo clean up. | |||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 03:37, 17 February 2010 (EST)== | ==[[User:Xiao Y. Liu|Xiao Y. Liu]] 03:37, 17 February 2010 (EST)== | ||
=== | ===sbb03 tos=== | ||
# set up PCA reaction for tos site. ( | # set up PCA reaction for tos site. (50ul reaction) | ||
====Assembly reaction==== | |||
* add following materials in order. ~ 5 min | |||
38 uL ddH2O | |||
5 ul 10x expand buffer | |||
5 ul 2mM dNTPs | |||
1 ul oligo mixture (100uM total, mixture of oligos after combination of 100uM stocks) | |||
0.75 ul Expand polymerase | |||
* run JCA/PCA1 program, (55°C for annealing) ~ 1.5 to 2 hours | |||
* stop here today. | |||
=== PiggyBac === | |||
====PCR of parts for SOEing==== | |||
# set up 3 PCR reactions for piggyBac. (33ul rxn) | |||
* basic PCR method to amplify your DNA from a plasmid or genomic DNA sample using the Expand polymerase. | * basic PCR method to amplify your DNA from a plasmid or genomic DNA sample using the Expand polymerase. | ||
| Line 207: | Line 284: | ||
* give GSI the tubes and run program. | * give GSI the tubes and run program. | ||
==[[User:Xiao Y. Liu|Xiao Y. Liu]] 12:51, 16 February 2010 (EST)== | ==[[User:Xiao Y. Liu|Xiao Y. Liu]] 12:51, 16 February 2010 (EST)== | ||
Latest revision as of 14:40, 21 April 2010
Xiao Y. Liu 17:40, 21 April 2010 (EDT)
- Team 1 project
- Link to team notebook SBB10AssayTeam1-LabNotebook
Xiao Y. Liu 16:06, 19 March 2010 (EDT)
PiggyBac
- miniprep and mapping
- stop at incubation at 37°C, Mike will run the gel.
Xiao Y. Liu 16:09, 17 March 2010 (EDT)
PiggyBac
- Gel Image of digested product
- zymo gel purification of digested product. elute with 8ul water.
- Ligation and Transformation.
Xiao Y. Liu 16:10, 10 March 2010 (EST)
PiggyBac
- obtained SOEing PCR product back, do an analytical gel(3ul pcr, 7ul loading buffer)
- zymo clean up. elute with 30ul water
Regular Zymo Cleanup
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction.
- Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction
- digestion
EcoRI/BamHI Digest of PCR Products
For PCR products, you will only digest a portion of your purified PCR product. Note that you must make a minor modification to the procedure if your DNA is shorter than 300bp (you add a little isopropanol to the ADB after melting).
- Set up the following reaction:
8uL of eluted PCR product 1uL of NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI
- Incubate at 37 degrees on the thermocycler for 1hr
- Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an acceptable stopping point
- If the DNA is shorter than 300bp, add 250uL of isopropanol and mix prior to loading it on the column
- dut to time manner, ask Will to do the gel and buffer for me.
sbb03-tos
- ligation and transformation
- obtained digested product from professor.
Ligation of EcoRI/BamHI digests
- Set up the following reaction:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest 1uL Insert digest 0.5uL T4 DNA Ligase
- Pound upside down on the bench to mix
- Give it a quick spin to send it back to the bottom of the tube
- Incubate on the benchtop for 30min
- Put on ice and proceed to the transformation
Transformation by heat-shock
Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock.
1. Thaw a 200 uL aliquot of cells on ice 2. Add 50 uL of water to the cells 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two 5. Add 70 uL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 90 seconds at 42 (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour 10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight
JCAnderson 18:59, 9 March 2010 (EST)

JCA reran the PCA using phusion instead of expand in step 1, came out much better (gel at right). Took through to digestion and cleanup, ready to ligate.
Xiao Y. Liu 16:16, 8 March 2010 (EST)
Xiao's gel on March 8th
Lane: 1. sbb03 2. PiggyBac 3. Ladder
PiggyBac
- after PCR 2, analytical gel (3ul PCR prodcut + 7ul loading buffer), look for 1813bp band
- Gel showed little SOEing products, so redo SOEing with 2k 45°C program.
SOEing
Set up the following reaction in a PCR tube:
24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo 1 = PiggyBac 01, 10uM 1uL Oligo 2 == PiggyBac 04, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA from A+B+C stock solution
Sbb03
- after Amplification, run a analytical gel (3ul PCR prodcut + 7ul loading buffer),look for 550bp band
- failed, so give all products to prof.
Xiao Y. Liu 16:12, 3 March 2010 (EST)
PiggyBac
- proceed SOEing PCR after zymo gel purifications for all 3 parts(A, B, C)
- Elute the DNA products from all 3 gel purification reactions in 50uL of water, since i eluted each one with 8.5 ul, after combining everything, add 50-3*8.5 = 24.5 ul dd water.
- Set up your second round of PCR as a normal 33uL reaction using the eluted mixture of fragments as template
Set up the following reaction in a PCR tube:
24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL Oligo 1 = PiggyBac 01, 10uM 1uL Oligo 2 == PiggyBac 04, 10uM 0.5uL Expand polymerase "1" 0.5uL Template DNA from A+B+C stock solution
sbb03
- after re-assembly, proceed to second round of amplification.
Amplification
Now, you need to do an amplification of the correct full-length chunks. Clean up the assembly reaction with a zymo column; don't bother running it on a gel - it'll be a smeary mess and won't really help you.
Regular Zymo Cleanup
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction.
- Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction (50ul in this case)
Save the purified product in case this step fails! For the amplification reaction, do a normal phusion program with 1 ul of the cleaned up assembly reaction as template, and using the outermost oligos for the chunk. That is:
Recipe
- 1 ul each outer oligo (10 uM)
- .5 ul phusion
- 10 ul 5x phusion buffer
- 5 ul 2mM dNTPs
- 32.5 ul H2O
Program
- 2 min initial denature at 94oC
- 30 sec denature at 94oC
- 30 sec anneal at 60oC [This should be high, as your outer oligos now have a huge overlap with the correct product]
- 30 sec extension at 68oC
- repeat 2-4 30 times total
Xiao Y. Liu 16:25, 1 March 2010 (EST)
PiggyBac
- Products from secomd PCR for piggyBac A, do a preparative gel with 6ul PCR products and 4ul loading buffer(blue, dye).
- prep gel showed success, so do a zymo gel purification for piggyBac A, eluted with 8.5ul water at the end. save stock and eluted DNA
sbb03
- Clean up the amplification reactions with a zymo column
Regular Zymo Cleanup
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction.
- Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction (50ul)
- mistake: forgot to do a analytic gel, so use 6ul of eluted product with 4ul loading buffer to run a analytical gel.
- gel showed nothing, so do a re-assembly with 1 ul PCR product.
Xiao Y. Liu 16:21, 24 February 2010 (EST)
PiggyBac
Second PCR trial for PiggyBac A
- PCR for piggyBac A failed, so do a second PCR with oligos(ca1530 and piggyBac02) and with program 55.
make dilution(10uM) puggyBac01 from stock(100uM) with 9ul water and 1ul stock
set up PCR tube:
- lable tubes sbb04 A Xiao Liu 2/23/10
24uL ddH2O
3.3uL 10x Expand Buffer "2"
3.3uL dNTPs (2mM in each)
1uL Oligo 1, 10uM
1uL Oligo 2, 10uM
0.5uL Template DNA
0.5uL Expand polymerase "1"
- place them in 55 PCR program.
PiggyBac B and PiggyBac C
- after preparative gel, zymo purification.
Zymo Gel Purification
- All spins until the drying step are 15 second full speed spins.
- transfer into the Zymo column inside a collection tube (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with 8.5 uL of water into a fresh Eppendorf tube
Sbb03
Amplification reaction with a normal phusion program
- contiune sbb03 experiment. PCR with tos-1 and tos-20
- add following in order: (check order with Prof. )
32.5 ul H2O 10 ul 5x phusion buffer 5 ul 2mM dNTPs 1 ul each outer oligo (10 uM)=tos-1 and tos-20 0.5 ul phusion
- run Phusion Program, anneal at 60°C,
Xiao Y. Liu 19:07, 22 February 2010 (EST)
PiggyBac
- Products from 3 PCR for piggyBac, do a preparative gel with sbb04A, sbb04B, and sbb04C with 6ul PCR products and 4ul loading buffer(blue, dye).
- sbb04 stops at melting gel with ADP buffer.
Image from preparative gel
1. ladder 2. sbb04A 3. sbb04B 4. sbb04C 5. RH 33A 6. RH33B 7. blank 8. blank 9. CD1 10. CD2 11. CD3 12. sbb19A 13. sbb19B 14. ladder
sbb03 tos
- After getting PCA products back for sbb03, do a zymo clean up.
Regular Zymo Cleanup
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction. It also will remove the buffer and restriction enzymes from a restriction digest reaction.
- Add 180 uL of Zymo ADB buffer (brown bottle) to a 33uL or 50uL reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube, use the same volume of water as the volume of the original reaction (500ul)
- sbb03 stops at the end of zymo clean up.
Xiao Y. Liu 03:37, 17 February 2010 (EST)
sbb03 tos
- set up PCA reaction for tos site. (50ul reaction)
Assembly reaction
- add following materials in order. ~ 5 min
38 uL ddH2O 5 ul 10x expand buffer 5 ul 2mM dNTPs 1 ul oligo mixture (100uM total, mixture of oligos after combination of 100uM stocks) 0.75 ul Expand polymerase
- run JCA/PCA1 program, (55°C for annealing) ~ 1.5 to 2 hours
- stop here today.
PiggyBac
PCR of parts for SOEing
- set up 3 PCR reactions for piggyBac. (33ul rxn)
- basic PCR method to amplify your DNA from a plasmid or genomic DNA sample using the Expand polymerase.
The oligo concentrations in my stocks should be 100uM. I use them at 10uM in this protocol. So, i first need to make an oligo dilution of:
9uL Water 1uL 100uM oligo
can throw away the remainder of the diluted oligo when i'm done, but hold onto the stock tube!
Set up the master mix in a PCR tube:
24uL ddH2O * 3 = 72 uL ddH2O
3.3uL 10x Expand Buffer "2" * 3 = 9.9uL 10x Expand Buffer "2"
3.3uL dNTPs (2mM in each) * 3 = 9.9 uL dNTPs
1.5uL Expand polymerase "1"
- lable tubes, divide the master mix into 3. and proceed following to each PCR reaction.
1uL Oligo 1, 10uM
1uL Oligo 2, 10uM
0.5uL Template DNA ( ask before this, Mike suggest to add this into master mix, but prof. said always add enzyme last, what should i do?)
- give GSI the tubes and run program.
Xiao Y. Liu 12:51, 16 February 2010 (EST)
- Planning for Experiment on Wed, Feb 17th, 2010
(sbb03) tos Site of N15 Flow Chart
- PCA with tos 1-tos 20, PCR with tos 1 and tos 20, 55°C.
- analytical gel to check length of products (550bp is the target sequence), cut out target band if find one.
- Zymo clean up
- digestion
- gel purify
- stop, check if enough time to proceed next step!!!
(sbb04) PiggyBac Transposase Flow Chart
- make master mix for 3 PCR reactions,
- PCR fragment A , B, and C
- analytic gel, check length
- Zymo clean up
- digestion, mapping?
- gel purify
- stop, check if enough time to proceed next step!!!
Xiao Y. Liu 18:30, 10 February 2010 (EST)
- check clotho, found that sbb04 DNA is not there, commit it.
- start tutorial 6
- correct mistakes in protein fusion construction files and ApE.
Xiao Y. Liu 18:26, 8 February 2010 (EST)
- modified construction file for both tos and piggyBac.
- upload sbb03 and sbb04 to clotho
- upload oligos to oligo log, and construction files.
Construction file for tos (modified)
1) sbb03: tos site of N15
Pool tos01 through tos20, assemble by PCA
PCR tos01/tos20 on PCA reaction (550bp,EcoR1/BamH1, gp)
Sub into pBca9145-jtk2625 (EcoRI/BamHI, 2057+929bp, L)
Product is pBca9145-sbb03 {<tos>}
------------
tos01 PCA assembly of tos CCATAGAATTCATGAGATCTTCTAAGCGCAACGGTATTACTTACGT
tos02 PCA assembly of tos ATTTAAAATCATTAAGTTAGGTTTTAAATATACCAACGTAAGTAATACCGTTGC
tos03 PCA assembly of tos TTAAAACCTAACTTAATGATTTTAAATGATAATAAATCATACCAATTGC
tos04 PCA assembly of tos CAGCATGTTCGCTTAACTTTTGATAGCAATTGGTATGATTTATTATC
tos05 PCA assembly of tos AAAGTTAAGCGAACATGCTGATTTTCACGCTGTTTATACACTTTGAG
tos06 PCA assembly of tos ATATAGAGACGGAAGAGATAGAGATGCCTCAAAGTGTATAAACAGCG
tos07 PCA assembly of tos TATCTCTTCCGTCTCTATATTGAAACACAATCAAAGAACATCAATCC
tos08 PCA assembly of tos TTCTTAGATAGTGGGGGATGTCACATGGATTGATGTTCTTTGATTG
tos09 PCA assembly of tos ATCCCCCACTATCTAAGAACACCATAACAGAACACAACATAGGAATG
tos10 PCA assembly of tos TGTTCCGAATTATTGATACATTAATGTTGCATTCCTATGTTGTGTTCTG
tos11 PCA assembly of tos AATGTATCAATAATTCGGAACATATGCACTATATCATATCTCAATTACGG
tos12 PCA assembly of tos ATGGGCAATTGTGTGCTGATATGTTCCGTAATTGAGATATGATATAG
tos13 PCA assembly of tos TCAGCACACAATTGCCCATTATACGCGCGTATAATGGACTATTGTGTG
tos14 PCA assembly of tos GTTCTGCGCTTATGTTCTCCTTATCAGCACACAATAGTCCATTATAC
tos15 PCA assembly of tos GAGAACATAAGCGCAGAACAATATGTATCTATTCCGGTGTTGTGTTC
tos16 PCA assembly of tos TATAAGAGAACATAATAGCAGAATAACAAAGGAACACAACACCGGAATAG
tos17 PCA assembly of tos TCTGCTATTATGTTCTCTTATAGTGTGACGAAAGCAGCATAATTAATC
tos18 PCA assembly of tos ATATCGTAACACAATCAAAGAACAAGTGACGATTAATTATGCTGCTTTCG
tos19 PCA assembly of tos TTCTTTGATTGTGTTACGATATCCAGAGACTTAGAAACGGGGGAAGG
tos20 PCA assembly of tos CATAGGGATCCTTCCCCCGTTTCTAAGT
Construction file for piggyBac transposase (modified)
Construction file for piggyBac transposase (sbb04)
PCR with piggyBac01/CA1530 on Bca1519 3-2 (335bp, gp=A)
PCR CA1527/piggyBac03 on Bca1519 3-1 (721bp, gp=B)
PCR with piggyBac02/piggyBac04 on Bca1553 4-6 (925bp, gp=C)
-----------------------------------------------------------------
PCR with piggyBac01 and piggyBac04 on A+B+C (1813bp, EcoRI/BamHI)
Digest pBjk2741-Bca1144 (EcoRI/BamHI, 2170bp + 910bp,L)
Product is pBjk2741-sbb04 {<piggyBac!}
-----------------------------------------------------------------
piggyBac01 Forward PCR of part 1 of piggyBac
ccatagaattcatgAGATCTGGTTGCTCTCTGGACGACGAAC
CA1527 TGGACGAACAGAACGTTATCGAACAGCCGGGTTCTTCTCTGGCTTCT
PCA assembly of piggiebacfiveprime (Bca1519)
CA1530 ACGACGGGTAGATTTAGAGGTAGACCAGCAGTGTTTGTTTTTACCAC
PCA assembly of piggiebacfiveprime (Bca1519)
piggyBac02 Forward SOEing part of piggyBac
ACGGTATCAAAATCCTGATGatgtgcgactctggtaccaa
piggyBac03 Reverse SOEing part of piggyBac
ttggtaccagagtcgcacatCATCAGGATTTTGATACCGT
piggyBac04 Reverse PCR of part 2 of piggyBac
gctagGGATCCttaGAAGCAAGACTGGCACATG
Xiao Y. Liu 15:50, 8 February 2010 (EST)
PiggyBac
- propose a construction file for sbb04, piggyBac transposase,
- question about whether to leave a start codon in the beginning of sequence. since the falimy flavor is {<part!}, so i leave it out.
2) construction file for piggyBac transposase (sbb04)
PCR with piggyBac01 and piggyBac03 on pBca9523-Bca1519 (928bp, gp, A)
PCR with piggyBac02 and piggyBac04 on pBca9523-Bca1553 (925bp, gp, B)
-----------------------------------------------------------------
PCR with piggyBac01 and piggyBac04 on A+B (1813bp, EcoR1/BamH1)
Digest pBjk2741-Bca1144 (EcoR1/BamH1, 2170bp + 910bp,L)
Product is pBjk2741-sbb04 {<piggyBac!}
-----------------------------------------------------------------
piggyBac01 Forward PCR of part 1 of piggyBac ccatagaattcatgAGATCTGGTTGCTCTCTGGACGACGAAC
piggyBac02 Forward SOEing part of piggyBac ACGGTATCAAAATCCTGATGatgtgcgactctggtaccaa
piggyBac03 Reverse SOEing part of piggyBac ttggtaccagagtcgcacatCATCAGGATTTTGATACCGT
piggyBac04 Reverse PCR of part 2 of piggyBac gctagGGATCCttaGAAGCAAGACTGGCACATG
- description of PCR product: ccata + EcoR1 + atg + bgl2 + part 1 (last 20bp + first 20bp as SOEing Part) and part 2 of piggyBac + stop codon + BamH1 + ctagc.
Xiao Y. Liu 20:00, 6 February 2010 (EST)
Sbb03 tos
oligo design for {<tos>} using geneDesign
- 1 biolding block, length of building block is 550 bp,
- average Tm of oligo overlaps in building block is 46°,
- number of oligos in building block: 20,
- average oligo length: 47bp, longest oligo: 54bp, shorest oligo: 28bp.
- PCA product is 5 random + EcoR1 + atg + bgl2 + target sequence + BamH1 + 5 random
Construction file for tos (check for errors)
1) sbb03: tos site of N15 Pool tos01 through tos20, assemble by PCA PCR tos01/tos20 on PCA reaction (550bp,EcoR1/BamH1, gp) Sub into pBca9145-jtk2625 (EcoRI/BamHI, 2057+929bp, L) Product is pBca9145-sbb03 ------------ tos01 PCA assembly of tos CCATAGAATTCATGAGATCTTCTAAGCGCAACGGTATTACTTACGT tos02 PCA assembly of tos ATTTAAAATCATTAAGTTAGGTTTTAAATATACCAACGTAAGTAATACCGTTGC tos03 PCA assembly of tos TTAAAACCTAACTTAATGATTTTAAATGATAATAAATCATACCAATTGC tos04 PCA assembly of tos CAGCATGTTCGCTTAACTTTTGATAGCAATTGGTATGATTTATTATC tos05 PCA assembly of tos AAAGTTAAGCGAACATGCTGATTTTCACGCTGTTTATACACTTTGAG tos06 PCA assembly of tos ATATAGAGACGGAAGAGATAGAGATGCCTCAAAGTGTATAAACAGCG tos07 PCA assembly of tos TATCTCTTCCGTCTCTATATTGAAACACAATCAAAGAACATCAATCC tos08 PCA assembly of tos TTCTTAGATAGTGGGGGATGTCACATGGATTGATGTTCTTTGATTG tos09 PCA assembly of tos ATCCCCCACTATCTAAGAACACCATAACAGAACACAACATAGGAATG tos10 PCA assembly of tos TGTTCCGAATTATTGATACATTAATGTTGCATTCCTATGTTGTGTTCTG tos11 PCA assembly of tos AATGTATCAATAATTCGGAACATATGCACTATATCATATCTCAATTACGG tos12 PCA assembly of tos ATGGGCAATTGTGTGCTGATATGTTCCGTAATTGAGATATGATATAG tos13 PCA assembly of tos TCAGCACACAATTGCCCATTATACGCGCGTATAATGGACTATTGTGTG tos14 PCA assembly of tos GTTCTGCGCTTATGTTCTCCTTATCAGCACACAATAGTCCATTATAC tos15 PCA assembly of tos GAGAACATAAGCGCAGAACAATATGTATCTATTCCGGTGTTGTGTTC tos16 PCA assembly of tos TATAAGAGAACATAATAGCAGAATAACAAAGGAACACAACACCGGAATAG tos17 PCA assembly of tos TCTGCTATTATGTTCTCTTATAGTGTGACGAAAGCAGCATAATTAATC tos18 PCA assembly of tos ATATCGTAACACAATCAAAGAACAAGTGACGATTAATTATGCTGCTTTCG tos19 PCA assembly of tos TTCTTTGATTGTGTTACGATATCCAGAGACTTAGAAACGGGGGAAGG tos20 PCA assembly of tos CATAGGGATCCTTCCCCCGTTTCTAAGT