SBB10Ntbk-WeiChunHsu
Part Synthesis (aka Wet Lab)
Wei-Chun Hu 18:18, 24 February 2010 (EST)
Protocol changes
- Change for protocol (replace with small fragment zymo cleanup) for PCA
- Did not get to Digest step (advised to hold off due to running time, and given possible failure of 4ABC...)
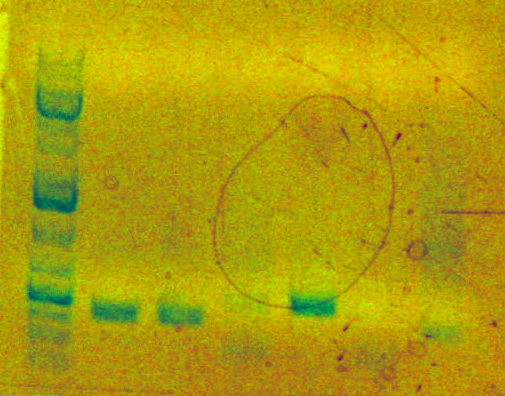
Products
- 4D - zymo-ed PCA (band looks fine, but faint)
- 5D - incomplete digest reaction - contains 8 uL of 4D
- 5ABC - zymo-ed SOEing PCR (problem: see below)
- 6ABC - incomplete digest reaction - contains 8 uL of 5ABC
Potential problems
- Band for 4ABC analytical gel is WAY off (5th from the left band is a bit over 500, product should be around 1800). Jeni also had the same problem with SOEing (band on far right). Recommended change SOEing protocol (SLIC, Venter?)
Wei-Chun Hu 15:31, 23 February 2010 (EST)
Protocol: PCA
- Optional: Run an analytical gel - 3 uL PCR, 7 uL loading buffer
- This gel is probably not necessary because we will do a gel in the digest step
- Band should be around 250 bp (below the 1st bright one)
- Clean up the amplification reactions with a zymo column
- Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction.
- Transfer into the Zymo column (small clear guys)
- Add 500uL of Ethanol and pipette up and down to mix
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube
- Digest the product and desired backbone with Eco/Bam:
- Set up the following reaction (two tubes):
- 8uL of eluted PCR product (backbone [pBjk2741-Bca1144] and PCA product in SEPARATE reactions)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Incubate at 37 degrees on the thermocycler for 1hr
- Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an acceptable stopping point
- Only proceed below if ample time
- Set up a Zymo column
- The DNA is shorter than 300bp: add 250uL of isopropanol and mix prior to loading it on the column
- Set up the following reaction (two tubes):
Protocol: SOE
- Optional: Run an analytical gel - 3 uL PCR, 7 uL loading buffer
- Expect bright band around 1800 bp (just above the middle bright one)
- Clean up the amplification reactions with a zymo column
- Add 180 uL of Zymo ADB buffer (brown bottle) to the reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube
- Digest the product and desired backbone with Eco/Bam:
- Set up the following reaction (we already have the plasmid digest):
- 8uL of eluted PCR product (PCR product)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Incubate at 37 degrees on the thermocycler for 1hr
- Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an acceptable stopping point
- Set up the following reaction (we already have the plasmid digest):
Wei-Chun Hu 17:31, 22 February 2010 (EST)
Results
Gel for PCR
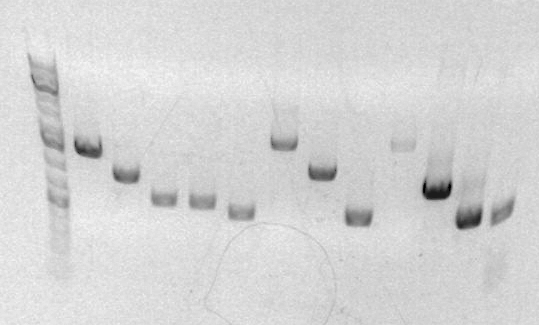
Last three columns are A,B,C. Lengths appear to be correct (~900,~500,~450). Distortion of gel makes it impossible to accurately compare the last two.
Protocol changes
8uL6uL of PCR product premixed with1uL4uL of loading buffer- Used Jeni's mastermix for 4ABC PCR because frankly everyone was running out of time
Products
- PCA
- 2D - zymo cleanup of 1D
- 3D - PCR amplification of 2D (put in PCA box)
- SOE
- JHA,JHB,JHC - preparative gels of 1A,1B,1C (6 uL PCR product / 4 uL 1X loading buffer)
- 2ABC - purifying gel fragments 1A,1B,1C
- 3ABC - purified 50uL DNA from gel fragments
- 4ABC - PCR reaction using 0.5 uL of 3ABC (put in 2K55 box)
Wei-Chun Hu 18:21, 21 February 2010 (EST)
Protocol: SOE
(This assumes the PCR has completed successfully.)
- For each PCR, load 6uL of PCR product premixed with 4uL of loading buffer in a single well of a 1% agarose gel
- Cut out the bands, put them into a single 1.5mL microcentrifuge tube (put ALL bands in one tube)
- Add 650uL of ADB Buffer
Zymo gel purification
- All spins until the drying step are 15 second full speed spins.
cut out bands minimizing extra gel matter.put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle).- heat at 55, shake and/or vortex until the gel has dissolved.
- (do NOT put isopropanol.)
- transfer into the Zymo column inside a collection tube (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- discard collection tube
- Elute the DNA in 50uL of water in a LABELED eppendorf tube
- Dilute 10 uM stocks of OJIMH011 and OJIMH014.
- Set up your second round of PCR as a normal 33uL reaction using the eluted mixture of fragments as template:
24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL OJIMH011, 10uM (dilute 100 uM stock) 1uL OJIMH014, 10uM (dilute 100 uM stock) 0.5uL Expand polymerase "1" 0.5uL Template DNA
Protocol: PCA
Small-Frag Zymo Cleanup
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction for fragments smaller than 300bp. It also will remove the buffer and restriction enzymes from a restriction digest reaction.
- Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction.
- Transfer into the Zymo column (small clear guys)
- Add 500uL of Ethanol and pipette up and down to mix
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube
Amplification
Now, you need to do an amplification of the correct full-length chunks. Clean up the assembly reaction with a zymo column; don't bother running it on a gel - it'll be a smeary mess and won't really help you. Save the purified product in case this step fails! For the amplification reaction, do a normal phusion program with 1 ul of the cleaned up assembly reaction as template, and using the outermost oligos for the chunk. That is:
Recipe
- 1 ul each outer oligo [sbb11 F/R] (10 uM)
- .5 ul phusion
- 10 ul 5x phusion buffer
- 5 ul 2mM dNTPs
- 32.5 ul H2O
Program
- 2 min initial denature at 94oC
- 30 sec denature at 94oC
- 30 sec anneal at 60oC [This should be high, as your outer oligos now have a huge overlap with the correct product]
- 30 sec extension at 68oC
- repeat 2-4 30 times total
Wei-Chun Hu 17:33, 17 February 2010 (EST)
Protocol
Resuspend lypophilized DNA. Oligo stock is 100 uM, dilute to 10 uM.
Appropriate components (see Media:2-17-10-cloning-PCR.pdf) were added to three Eppendorf tubes. Transfer to PCR tubes at end. DNA polymerase was added to the mastermix.
Oligos for PCA were provided as mixture (100 uM, M), forward (10 uM, F), and reverse (10 uM, R). The Template:SBB-PCA protocol was used.
Products
1A: OJIMH011,OJIMH013,Bca1623 6-9
1B: OJIMH012,CA1674,Bca1559 7-7
1C: CA1675,OJIMH014,Bca1559 7-1
1D: Oligo mixture (M SBB11)
Wei-Chun Hu 18:10, 16 February 2010 (EST)
Run this protocol tomorrow: Media:2-17-10-cloning-PCR.pdf
Wei-Chun Hu 14:24, 9 February 2010 (EST)
The following protocols are relevant for me:
Cloning by PCR
SOEing PCR
PCA Gene Synthesis
Regular Zymo Cleanup
Zymo Gel Purification
EcoRI/BamHI Digest of PCR Products
Ligation of EcoRI/BamHI digests
Transformation by heat-shock
Picking of colonies
Miniprep purification of DNA
Computational Part Design
Wei-Chun Hu 01:45, 4 February 2010 (EST)
All construction files (now transcluded)
1) sbb11: sleeping beauty 5'TR Pool OJIMH001 through OJIMH010, assemble by PCA PCR OJIMH001/OJIMH010 on PCA reaction (257 bp, EcoRI/BamHI) Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L) Product is pBjk2741-sbb11 ------------ OJIMH001 PCA assembly of sbb11 CCATAGAATTCATGAGATCTAGTTGAAGTCGGAAGTTTACATACACTTAAG OJIMH002 PCA assembly of sbb11 GAAAAACGAGTTTTAATGACTCCAACTTAAGTGTATGTAAACTTCCGACTTC OJIMH003 PCA assembly of sbb11 AAGTTGGAGTCATTAAAACTCGTTTTTCAACTACACCACAAATTTCTTGTTAAC OJIMH004 PCA assembly of sbb11 CTGACTTGCCAAAACTATTGTTTGTTAACAAGAAATTTGTGGTGTAGTTGAAA OJIMH005 PCA assembly of sbb11 TTAACAAACAATAGTTTTGGCAAGTCAGTTAGGACATCTACTTTGTGC OJIMH006 PCA assembly of sbb11 ACAATTGTTGGAAAAATGACTTGTGTCATGCACAAAGTAGATGTCCTAACTG OJIMH007 PCA assembly of sbb11 GACACAAGTCATTTTTCCAACAATTGTTTACAGACAGATTATTTCACTTATAATTC OJIMH008 PCA assembly of sbb11 CCACTGGAATTGTGATACAGTGAATTATAAGTGAAATAATCTGTCTGTAAAC OJIMH009 PCA assembly of sbb11 TAATTCACTGTATCACAATTCCAGTGGGTCAGAAGTTTACATACACTAAG OJIMH010 PCA assembly of sbb11 CTGATGGATCCTTAGTGTATGTAAACTTCTGACCC
JCA Notes
- Correct
2) sbb13: PhiC31 Integrase
PCR OJIMH011/OJIMH012 on pBca9523-Bca1623 (949 bp, gp = A)
PCR OJIMH013/OJIMH014 on pBca9523-Bca1659 (938 bp, gp = B)
-------------
PCR PJIMH011/OJIMH014 on A+B (1843 bp, EcoRI/BamHI)
Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L)
Product is pBjk2741-sbb13 {<phiC31>}
-------------
OJIMH011 Forward PCR of Part 1 of PhiC31 ccataGAATTCatgAGATCTGACACGTACGCGGGTGCTTA
OJIMH012 SOEing of Part 1 (R) of PhiC31 AAGAAGCCGGACGGCACGCCGACCacgaagattgagggttaccg
OJIMH013 SOEing of Part 2 (F) of PhiC31 cggtaaccctcaatcttcgtGGTCGGCGTGCCGTCCGGCTTCTT
OJIMH014 Reverse PCR of Part 2 of PhiC31 atcagGGATCCCGCCGCTACGTCTTCCGT
JCA Notes
- Oligos 12 and 13 are switched
- Otherwise correct
sbb13: PhiC31 Integrase PCR OJIMH011/OJIMH013 on Bca1623 6-9 (949bp, gp=A) PCR OJIMH012/CA1674 on Bca1559 7-7 (514bp, gp=B) PCR CA1675/OJIMH014 on Bca1559 7-1 (444bp, gp = C) ------------- PCR PJIMH011/OJIMH014 on A+B+C (1843 bp, EcoRI/BamHI) Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L) Product is pBjk2741-sbb13 {<phiC31>} ------------- OJIMH011 Forward PCR of Part 1 of PhiC31 ccataGAATTCatgAGATCTGACACGTACGCGGGTGCTTA OJIMH012 SOEing of Part 1 (R) of PhiC31 AAGAAGCCGGACGGCACGCCGACCacgaagattgagggttaccg OJIMH013 SOEing of Part 2 (F) of PhiC31 cggtaaccctcaatcttcgtGGTCGGCGTGCCGTCCGGCTTCTT CA1674 GGGCGTCGGCGCGCTCCGCAACAAGGTTCGCCCGTTCGCCGCTCTTCTCAG PCA assembly of phiCthreeprime (Bca1659) CA1675 TGCGGAGCGCGCCGACGCCCTGAACGCCCTTGAAGAGCTGTACGAAGACCG PCA assembly of phiCthreeprime (Bca1659) OJIMH014 Reverse PCR of Part 2 of PhiC31 atcagGGATCCCGCCGCTACGTCTTCCGT
Wei-Chun Hu 20:45, 3 February 2010 (EST)
Construction file for sbb13 (this one is especially tricky, so please check)
2) sbb13: PhiC31 Integrase
PCR OJIMH011/OJIMH012 on pBca9523-Bca1623 (949 bp, gp = A)
PCR OJIMH013/OJIMH014 on pBca9523-Bca1659 (938 bp, gp = B)
-------------
PCR PJIMH011/OJIM014 on A+B (1843 bp, EcoRI/BamHI)
Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L)
Product is pBjk2741-sbb13 {<phiC31>}
-------------
OJIMH011 Forward PCR of Part 1 of PhiC31 ccataGAATTCatgAGATCTGACACGTACGCGGGTGCTTA
OJIMH012 SOEing of Part 1 (R) of PhiC31 AAGAAGCCGGACGGCACGCCGACCacgaagattgagggttaccg
OJIMH013 SOEing of Part 2 (F) of PhiC31 cggtaaccctcaatcttcgtGGTCGGCGTGCCGTCCGGCTTCTT
OJIMH014 Reverse PCR of Part 2 of PhiC31 atcagGGATCCCGCCGCTACGTCTTCCGT
Wei-Chun Hu 20:03, 3 February 2010 (EST)
Construction file for sbb11 (check for errors)
1) sbb11: sleeping beauty 5'TR Pool OJIMH001 through OJIMH010, assemble by PCA PCR OJIMH001/OJIMH010 on PCA reaction (257 bp, EcoRI/BamHI) Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L) Product is pBjk2741-sbb11 ------------ OJIMH001 PCA assembly of sbb11 CCATAGAATTCATGAGATCTAGTTGAAGTCGGAAGTTTACATACACTTAAG OJIMH002 PCA assembly of sbb11 GAAAAACGAGTTTTAATGACTCCAACTTAAGTGTATGTAAACTTCCGACTTC OJIMH003 PCA assembly of sbb11 AAGTTGGAGTCATTAAAACTCGTTTTTCAACTACACCACAAATTTCTTGTTAAC OJIMH004 PCA assembly of sbb11 CTGACTTGCCAAAACTATTGTTTGTTAACAAGAAATTTGTGGTGTAGTTGAAA OJIMH005 PCA assembly of sbb11 TTAACAAACAATAGTTTTGGCAAGTCAGTTAGGACATCTACTTTGTGC OJIMH006 PCA assembly of sbb11 ACAATTGTTGGAAAAATGACTTGTGTCATGCACAAAGTAGATGTCCTAACTG OJIMH007 PCA assembly of sbb11 GACACAAGTCATTTTTCCAACAATTGTTTACAGACAGATTATTTCACTTATAATTC OJIMH008 PCA assembly of sbb11 CCACTGGAATTGTGATACAGTGAATTATAAGTGAAATAATCTGTCTGTAAAC OJIMH009 PCA assembly of sbb11 TAATTCACTGTATCACAATTCCAGTGGGTCAGAAGTTTACATACACTAAG OJIMH010 PCA assembly of sbb11 CTGATGGATCCTTAGTGTATGTAAACTTCTGACCC
Wei-Chun Hu 19:45, 3 February 2010 (EST)
A test entry.