SBB10Ntbk-WeiChunHsu: Difference between revisions
Wei-Chun Hu (talk | contribs) (→PCA) |
Wei-Chun Hu (talk | contribs) No edit summary |
||
| (33 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__TOC__ | __TOC__ | ||
=Assays= | |||
[[Media:elisa-data.xls]] | |||
==4/28/2010== | |||
[[Media:elisa-data.xls]]<br> | |||
{| | |||
|[[Image:Absorbance.jpg|thumb|300px|Absorbance vs. concentration of T4 Ligase]] | |||
|[[Image:Absorbance of tagged T4 ligase.jpg|thumb|280px|Absorbance vs. T4 Ligase with various tags]] | |||
|} | |||
===ELISA Results=== | |||
The graph at the right displays a basic breakdown of the absorbance data acquired at the end of the ELISA assay. The four columns indicate: | |||
#No T4L - T4 ligase was not added to these wells, meaning there was nothing for the antibodies to bind in the assay. | |||
#No Myc - The Myc antibody was not added to these wells, meaning there was nothing to bind the T4 ligase to the plate. | |||
#No his-HRP - The his antibody conjugated to HRP was not added to these wells, meaning that the T4 ligase was not attached to a second antibody, and also that no reaction would occur with the HRP to produce a absorbance signal. | |||
#Pos - This is the positive control, so all reagents should have been added at the appropriate parts of the protocol. It should produce the largest absorbance signal if the T4 ligase does contain both myc and his tags and was successfully bound. | |||
From the values of each bar, we can get some information about the effectiveness and background absorbance of this assay. <br> | |||
<br> | |||
Ideally, the 'no his-HRP' control should display baseline absorbance because HRP is the molecule responsible for oxidizing TMB and turn blue which has highest absorbance in 450nm, indicating the existence of T4 Ligase. regardless to whether HRP and TMB exist, there are some chemicals in the solution that display some background absorbance. so this should be treated as a loose baseline to compare the rest of the data to. <br> | |||
The 'no myc' and 'no T4L' controls both produced similar levels of fluorescence to each other. This is consistent with the idea that in both of these controls, there should be no T4L bound to the plate (either because it was not present or had nothing to bind to). Even so, it seems that the his-HRP, even without T4L, was able to bind in some amount to the plate and increase the absorbance signal as compared to the no his-HRP case. It is possible that his would be able to interact with the myc antibody or attach to the plate even with the blocking buffer present. These controls indicate that the absorbance signal must be higher than about .2 absorbance to effectively indicate a working ELISA assay. <br> | |||
In fact, the 'pos' control gives a absorbance signal of about .27 absorbance, the highest of the four controls. This may indicate that the T4 ligase was effectively bound and measured by the two antibodies. However, the standard deviation from this control is so large that it overlaps with the 'no myc' and 'no T4L' controls. This variation may be due to the differing concentrations of T4 ligase which were added to the wells, or other factors which would require repeats of this experiment. | |||
===Suggestions for ELISA=== | |||
1. '''Problem: Absorbances remain unchanged/increases with decreasing protein concentration''' | |||
<blockquote> | |||
Possible cause: Protein concentrates saturates available antibody<br> | |||
Solution 1: Reduce protein concentrations below 1/10000X<br> | |||
Solution 2: Perform a second purification step<br> | |||
Solution 3: Increase antibody concentrations available to bind protein (primary detecting (anti-His) or primary capture (anti-Myc))<br> | |||
</blockquote> | |||
<p> | |||
2. '''Problem: Absorbance of positive control is not significantly different from negative controls''' | |||
<blockquote> | |||
Possible cause: steric hinderance of dual tag construct. <br> | |||
Solution 1: add a ALA-6 linker between two tags. <br> | |||
Solution 2: put tags on the opposite side of the protein. <br> | |||
second possible cause: background noise. <br> | |||
solution 1: do more replicates. <br> | |||
</blockquote> | |||
===TECAN data=== | |||
[[Media:TECAN-spreadsheet.xls]]<br> | |||
====Legend==== | |||
* X axis = Cycles (each cycle is approximately 5 minutes) | |||
* Y axis = Absorbance (relative units, at 600 nm) | |||
* <font color=blue>A,F (blue) = GP2578</font> | |||
* <font color=red>B,G (red) = GP2599</font> | |||
* <font color=green>C,H (green) = GP2619</font> | |||
====Without arabinose induction==== | |||
{| | |||
|[[Image:te-1no.png|thumb|250px|Replicate 1]] | |||
|[[Image:te-3no.png|thumb|250px|Replicate 2]] | |||
|[[Image:te-5no.png|thumb|250px|Replicate 3]] | |||
|} | |||
====With arabinose induction==== | |||
{| | |||
|[[Image:te-1yes.png|thumb|250px|Replicate 1]] | |||
|[[Image:te-3yes.png|thumb|250px|Replicate 2]] | |||
|[[Image:te-5yes.png|thumb|250px|Replicate 3]] | |||
|} | |||
===TECAN Analysis=== | |||
Cells did not show differential growth rates with arabinose induced constructs, as expected. No growth was observed for many wells that are near the edges of the TECAN plate; recommend to not use edge wells in future analyses. | |||
==4/26/2010== | |||
Did ELISA. Left lab around 5 pm; last step performed was incubating 1h after adding varying concentrations of T4 ligase to 24 wells. | |||
<br><br> | |||
Protocol (roughly):<br> | |||
# Dilute anti-Myc antibody (1000X) to 1X in TBST, plate 100 uL into well | |||
# Incubate at RT for 1h | |||
# Do three plate washes (TBST with 0.1% Tween) | |||
# Put blocking buffer (2% BSA, in TBST) 100 uL in wells, incubate 1h at RT | |||
# Do three plate washes | |||
# Put T4 Ligase at various concentrations (diluted in blocking buffer) (see attached spreadsheet) into wells | |||
# Incubate 1h at 37C | |||
# Do three washes | |||
# Put anti-His antibody conjugated to HRP (10000X) 100uL, dilute to 1X in blocking buffer, 1h at 37C | |||
# Do three washes | |||
# Put detection 3,3’,5,5’-Tetramethylbenzidine (TMB) solution 100uL | |||
# Incubate at 37C for 15 min | |||
# Detect with spectrometer at 450 nm | |||
==4/22/2010== | |||
# Get column | |||
# Pour in lysate + beads | |||
# Collect drips | |||
# Wash with ice cold PBS (20 mL) (N x 0.5 mL) | |||
# Elute with 4-5 1 mL volumes of 300 mM imidazole in PBS | |||
# Put eluant into dialysis column, add ice cold PBS to column, then shake in a speed of 5000mph with 4°C for 30 min, then change to fresh PBS, repeat shaking and change media for several times. | |||
# add ligase, purify and run in gel (Mike did this). | |||
==4/21/2010== | |||
[[Image:Notes-protein-purification.jpg|thumb|500 px|Notes for 4/21/2010]] | |||
* General protocol for protein purification: | |||
# Pick starter colony | |||
# Grow starter culture | |||
# Put in flask | |||
# Grow to midlog | |||
# Induce arabinose | |||
# Grow to saturation, have protein | |||
# '''Transfer to centrifuge tube (100 mL) x 2 - stuff here on is on ice (4C)''' | |||
# Spin 5 min at 6000 rpm | |||
# Dump supernatant, leave pellet | |||
# 10 mL of resuspension buffer (TBS), resuspend and combine in 50 mL conical flask | |||
# Sonicate (probe) - 1 minute (is it a program?). DO pulses. | |||
# Spin to remove debris | |||
# Transfer supernatant to fresh tube (pour). Will be translucent. | |||
# Add 400 uL of Ni resin | |||
# Agitate at 4C 1h to overnight | |||
* Tomorrow | |||
# Get column | |||
# Pour in lysate + beads | |||
# Collect drips | |||
# Wash with PBS (20 mL) (4 x 5 mL) | |||
# Elute with 4-5 1 mL volumes of 300 mM imidazole in PBS | |||
# Put eluant into dialysis bag | |||
# Let dialyze overnight on ice (4°C) | |||
==4/19/2010== | |||
* analyze TECAN data, maximize TECAN protocol | |||
** TECAN data showed significant edge effects. Recommended to not use outer rows and columns | |||
** I have no clue how to derive the actual growth rate. Tried some logistic equations (a*b)/(a-(b-a)*Exp(-c*t)) and nonlinear regression but my system keeps blowing up, regression looks bad, etc. | |||
** There are a lot of forms of logistic equations you can use. E.x. K / (1 + exp(a + b*x), the differential form dN/dt = r*N(K-N)/K, etc. If anyone has a copy of Mathematica or SPSS, try to play around with it. | |||
* develop ELISA protocol | |||
==4/15/2010== | |||
* materials: 20% 100X arabinose, LB media and 96 well plate for TECAN | |||
* dilute arabinose to 1X, 50ul LB in each well. 1ul of saturated cell mixture. | |||
* Plate: http://spreadsheets.google.com/ccc?key=0AvbXytCFRyFCdGJNVXhzNTUtWTgtZFlkYmZGTVQxT2c&hl=en | |||
* by accident, added two uls of different cells to F3 | |||
===TECAN=== | |||
[[Image:Tecansafireii-pic.jpg|thumb|upright|A TECAN Safire II machine at Architecture et Fonction des Macromolécules Biologiques]] | |||
'''Running a TECAN analysis:'''<br> | |||
''The procedure below only works for black 96-well flat bottom plates.''<br> | |||
#Turn TECAN power on (wait for light to stop flashing) | |||
#Open XFluor4 Safire II XLS spreadsheet | |||
#Load plate: | |||
##Have 96-well plate with media+cells | |||
##Goto XFluorSafireII menu > movements > out | |||
##Load plate | |||
##Goto XFluorSafireII menu > movements > in | |||
#Load the program to run: | |||
##Select Multi Labeling Kinetic | |||
##Load multi labeling kinetic parameter > "\iGEM 2007\My Documents\Weston\gabe's experimental folder\kinteticmodified_no gfp reading.mps" | |||
## Click "Run" | |||
# Wait for data collection. Operation can only be cancelled when machine is performing measurements. | |||
==4/14/2010== | |||
Picked 3 colonies from the 3 different constructs. Transformation efficiency is, of course, insane because there was no ligation performed. | |||
{| | |||
| [[Image:jtk2559-jtk2164-041410.jpg|thumb|upright|jtk2559 (2599?)]] | |||
| [[Image:jtk2578-jtk2164-041410.jpg|thumb|upright|jtk2578]] | |||
| [[Image:jtk2619-jtk2164-041410.jpg|thumb|upright|jtk2619]] | |||
|} | |||
Ugh, image markup is confusing. Anyways, refer to the edit page for code to do the above. | |||
==4/12/2010== | |||
*Trial run for transformation. Obtained vectors. | |||
# jtk2559-jtk2164 K | |||
# jtk2619-jtk2164 K | |||
# jtk2578-jtk2164 K | |||
* Transformation protocol: | |||
# Add: | |||
## plasmid 0.5 uL | |||
## cells (MC1061 pir+) 10 uL | |||
# Add to cells: | |||
## KCM 1.5 uL | |||
## Water 2.5 uL | |||
# Use normal heat shock | |||
## Did @ 42C for 120 sec | |||
# Rescue (rescued 40 min) | |||
# Plate | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 19:36, 31 March 2010 (EDT)== | |||
Our team page: [[Template:SBB10AssayTeam1]]<br> | |||
Team notes: [[SBB10AssayTeam1-Notes]] | |||
=Part Synthesis (aka Wet Lab)= | =Part Synthesis (aka Wet Lab)= | ||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 23:47, 17 March 2010 (EDT)== | |||
Chose sbb13-1 and sbb13-2 for sequencing. Put into clotho. | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 00:42, 17 March 2010 (EDT)== | |||
===Protocol=== | |||
<S>Awaiting sequence results. Repeat Miniprep if needed.</s> Redo sbb13 (as I predicted), depending on what JCA says. | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 19:27, 15 March 2010 (EDT)== | |||
===Products=== | |||
*sbb11-1, sbb11-2, sbb11-3, sbb11-4 - 4 clones picked from sbb11 plate | |||
*sbb13-1, sbb13-2, sbb13-3, sbb13-4 - 4 clones picked from sbb13 plate | |||
[[Image:WJH_plates_031510.jpg]] | |||
*Very (VERY) rough estimation of transformation efficiency: | |||
**sbb11 - (87*17 cells)/(((50 ng))*(1 uL/8.5uL)) = 251 cells/ng insert = 23 cells/ng plasmid | |||
**sbb13 - (42*17 cells)/(((80 ng)*2)*(1 uL/8.5uL)) = 38 cells/ng insert = 17 cells/ng plasmid | |||
**sbb11 transformed nearly 10x as efficiently (gramwise), and approximately the same efficiency molewise (1812/226). | |||
*Choose: sbb11-2, sbb13-4 for sequencing | |||
[[Image:WJH_gels_031510.jpg]] | |||
*Lanes 2-5 look fine (Expected: 1727/684 (EcoRI/XhoI), actual ~1700/650) - chose sbb11-2 (lane 3) | |||
*Lanes 10-13 - (Expected: 2170/1827 (EcoRI/BamHI), actual (sbb13-4): ~2500/1750) - need more resolution to be sure. Chose sbb13-4. If it fails, choose sbb13-1 or sbb13-2. | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 03:07, 15 March 2010 (EDT)== | |||
===Protocol=== | |||
''Assume white colonies are present. This may or may not be a reasonable assumption.'' | |||
====Miniprep purification of DNA using Macherey-Nagel Nucleospin Kit==== | |||
'''MINIPREP (1mL - 5mL) Procedure for Plasmid DNA Purification''' | |||
*Pellet the cells (for individual cultures) | |||
#Pellet around 1.5 mL to 3 mL saturated culture by spinning full speed, 30 seconds. | |||
#Dump supernatant, repeat to pellet another 1.5 mL (for a total of 3 mL if desired) | |||
*Pellet the cells (for 24-well blocks) | |||
#In 327, weigh the block on the scale, fill a dirty block with water to the same weight | |||
#Use the Allegra centrifuge to spin at 5500rpm for 4 minutes | |||
#Dump supernatant | |||
#You will need to transfer the cells to microcentrifuge tubes after resuspension in A1 in the alkaline lysis section below | |||
*Do an alkaline lysis | |||
#Add 250uL of A1 buffer into each tube. Resuspend the cells using a vortexer. | |||
#Add 250uL of A2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down. Solution should become clearer. | |||
#Add 300uL of A3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then shake. | |||
#Spin in centrifuge at top speed for 5 minutes. | |||
#Label columns with an alcohol-resistant lab pen. | |||
*Purify DNA on a column | |||
#Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds. | |||
#Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin) | |||
#Wash each column with 500 uL of AW buffer. | |||
#Spin in centrifuge at full speed for approximately 15 seconds, then flick out the liquid again. | |||
#Wash with 750uL of A4 buffer (washes the salts off the resins). | |||
#Spin in centrifuge at full speed for approximately 15 seconds and flick out liquid again. | |||
#Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol. | |||
#Label new tubes and put columns in them. | |||
#Elute them by squirting 50uL of water down the middle of the column (don't let it stick to the sides). | |||
#Spin in centrifuge at top speed for 30 seconds. | |||
#Take out columns and cap the tubes. | |||
#Clean up - note the A1 buffer is stored at 4degC and all the rest at room temperature. | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 14:07, 11 March 2010 (EST)== | |||
===Products=== | |||
#10D - transformed cells from 9D ligation rxn with pBjk2741 | |||
#8ABC - transformed cells from 7ABC ligation rxn with pBjk2741 | |||
''I don't know how well transformed cells keep under freezing. They are in box C regardless.'' | |||
===Notes=== | |||
*Strange "spots" noted on Spec+ plates. Could be contamination, but most likely air bubbles. | |||
*2YT media from last lab was contaminated. Samples should not be affected. | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 18:02, 9 March 2010 (EST)== | |||
===Protocol=== | |||
''Protocols from here on out should be the same for both procedures, excepting miniprep digest mapping.'' | |||
====Ligation==== | |||
*Set up the following reaction: | |||
6.5uL ddH2O | |||
1uL T4 DNA Ligase Buffer (small red or black-striped tubes) | |||
1uL Vector digest (pBjk2741-Bca1144) | |||
1uL Insert digest (9D or 7ABC) | |||
0.5uL T4 DNA Ligase | |||
*Pound upside down on the bench to mix | |||
*Give it a quick spin to send it back to the bottom of the tube | |||
*Incubate on the benchtop for 30min | |||
*Put on ice and proceed to the transformation | |||
====Transformation by heat-shock==== | |||
'''Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock.''' | |||
1. Thaw a 200 uL aliquot of cells on ice | |||
2. Add 50 uL of water to the cells | |||
3. Add 30 uL of KCM to the cells | |||
4. Put your ligation mixture on ice, let cool a minute or two | |||
5. Add 70 uL of the cell cocktail to the ligation, stir to mix | |||
6. Let sit on ice for 10 min | |||
7. Heat shock for 90 seconds at 42 (longer incubation may work better) | |||
8. Put back on ice for 1 min | |||
9. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour | |||
10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 19:14, 8 March 2010 (EST)== | |||
===Products=== | |||
#9D - Zymo prep gel of digest 7D1 | |||
#7ABC - Zymo prep gel of digest 5ABC2 | |||
[[Image:JKgel.jpg]] | |||
*Lane 2: Faint, ~250bp | |||
*Lane 3: Strong, ~1800bp | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 19:59, 7 March 2010 (EST)== | |||
===Protocol: PCA=== | |||
#Digest the product <b>(7D1)</b> and desired backbone with Eco/Bam: | |||
##Set up the following reaction (two tubes): | |||
###8uL of eluted PCR product (backbone [pBjk2741-Bca1144] and PCA product in SEPARATE reactions) | |||
###1uL of NEB Buffer 2 | |||
###0.5uL EcoRI | |||
###0.5uL BamHI | |||
# Incubate 1h | |||
# Load prep. gel | |||
# Zymo g.p. | |||
##cut out bands minimizing extra gel matter. | |||
##put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle). | |||
##heat at 55, shake and/or vortex until the gel has dissolved. | |||
##'''If the DNA is <300bp''' add 250uL of isopropanol | |||
##transfer into the Zymo column inside a collection tube (small clear guys) | |||
##spin through, discard waste. | |||
##Add 200 uL of PE buffer (which is basically 70% ethanol) | |||
##spin through, discard waste. | |||
##Add 200 uL of PE buffer | |||
##spin through, discard waste. | |||
##spin for 90 seconds, full speed to dry. | |||
##elute with 8.5 uL of water into a fresh Eppendorf tube | |||
===Protocol: SOE=== | |||
#Digest the product <B>(5ABC2)</b> and desired backbone with Eco/Bam: | |||
##Set up the following reaction (two tubes): | |||
###8uL of eluted PCR product (backbone [pBjk2741-Bca1144] and PCA product in SEPARATE reactions) | |||
###1uL of NEB Buffer 2 | |||
###0.5uL EcoRI | |||
###0.5uL BamHI | |||
# Incubate 1h | |||
# Load prep. gel | |||
# Zymo g.p. | |||
##cut out bands minimizing extra gel matter. | |||
##put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle). | |||
##heat at 55, shake and/or vortex until the gel has dissolved. | |||
##transfer into the Zymo column inside a collection tube (small clear guys) | |||
##spin through, discard waste. | |||
##Add 200 uL of PE buffer (which is basically 70% ethanol) | |||
##spin through, discard waste. | |||
##Add 200 uL of PE buffer | |||
##spin through, discard waste. | |||
##spin for 90 seconds, full speed to dry. | |||
##elute with 8.5 uL of water into a fresh Eppendorf tube | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 13:15, 4 March 2010 (EST)== | |||
===Overall schedule outline=== | |||
* Monday | |||
** Digest Eco/Bam (1h) | |||
** Prep. gel | |||
** Zymo gel purification | |||
* Wednesday | |||
** Ligate part and vector digests (30 min) | |||
** Transformation (rescue 1h) | |||
** Plate | |||
* Weekend | |||
** (GSI) Pick colonies | |||
* Next Monday | |||
** Miniprep | |||
** Analytical gel | |||
** Sequence | |||
** Done! | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 17:15, 3 March 2010 (EST)== | |||
===Products Zymo-ed=== | |||
''Chosen products <b>bolded</b>'' | |||
# <b>7D1 - Zymo of JHD (7D)</b> | |||
# 5D1 - Zymo of JHD2 (5D) | |||
# <b>5ABC2 - Zymo of H2O (4ABC2)</b> | |||
# 5ABC3 - Zymo of DMSO (4ABC3) | |||
===Gels=== | |||
[[Image:3-3-10-gel.png]]<br> | |||
#LADDER | |||
#sbb22 H2O | |||
#sbb22 DMSO | |||
#sbb20 H2O | |||
#sbb20 DMSO | |||
#sbb7 1/2 DMSO | |||
#sbb7 1/2 | |||
#JH7D | |||
#JH5D | |||
#JH4ABC2 | |||
#JH4ABC3 | |||
#AS123 | |||
#LADDER | |||
* Band for 7D and 5D look identical. I will choose 7D regardless because it has been purified more. | |||
* Band for 4ABC2 (H2O) and 4ABC3 (DMSO) are nearly identical except for some residual lower bp products in both. H2O looks slightly better than DMSO; however, both are usable. | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 04:16, 2 March 2010 (EST)== | |||
===A public service announcement=== | |||
Jim will be gone early on Wed and Fri for personal matters. | |||
==="Suggested" Protocol=== | |||
* Immediately set up 4 analytical gels for all PCRs. | |||
** Select the better out of (4ABC2 [h2o] or 4ABC3 [DMSO]) and (JHD [7D] or JHD2 [5D]). | |||
** Try to get the gels run ASAP. I really don't have much time. | |||
* If at least 2 out of 4 are satisfactory, do a zymo cleanup (if I leave at 3, that's probably as far as I will get.) | |||
==[[User:Wei-Chun Hu|Wei-Chun Hu]] 16:38, 1 March 2010 (EST)== | ==[[User:Wei-Chun Hu|Wei-Chun Hu]] 16:38, 1 March 2010 (EST)== | ||
Latest revision as of 11:30, 29 April 2010
Assays
4/28/2010
 |
 |
ELISA Results
The graph at the right displays a basic breakdown of the absorbance data acquired at the end of the ELISA assay. The four columns indicate:
- No T4L - T4 ligase was not added to these wells, meaning there was nothing for the antibodies to bind in the assay.
- No Myc - The Myc antibody was not added to these wells, meaning there was nothing to bind the T4 ligase to the plate.
- No his-HRP - The his antibody conjugated to HRP was not added to these wells, meaning that the T4 ligase was not attached to a second antibody, and also that no reaction would occur with the HRP to produce a absorbance signal.
- Pos - This is the positive control, so all reagents should have been added at the appropriate parts of the protocol. It should produce the largest absorbance signal if the T4 ligase does contain both myc and his tags and was successfully bound.
From the values of each bar, we can get some information about the effectiveness and background absorbance of this assay.
Ideally, the 'no his-HRP' control should display baseline absorbance because HRP is the molecule responsible for oxidizing TMB and turn blue which has highest absorbance in 450nm, indicating the existence of T4 Ligase. regardless to whether HRP and TMB exist, there are some chemicals in the solution that display some background absorbance. so this should be treated as a loose baseline to compare the rest of the data to.
The 'no myc' and 'no T4L' controls both produced similar levels of fluorescence to each other. This is consistent with the idea that in both of these controls, there should be no T4L bound to the plate (either because it was not present or had nothing to bind to). Even so, it seems that the his-HRP, even without T4L, was able to bind in some amount to the plate and increase the absorbance signal as compared to the no his-HRP case. It is possible that his would be able to interact with the myc antibody or attach to the plate even with the blocking buffer present. These controls indicate that the absorbance signal must be higher than about .2 absorbance to effectively indicate a working ELISA assay.
In fact, the 'pos' control gives a absorbance signal of about .27 absorbance, the highest of the four controls. This may indicate that the T4 ligase was effectively bound and measured by the two antibodies. However, the standard deviation from this control is so large that it overlaps with the 'no myc' and 'no T4L' controls. This variation may be due to the differing concentrations of T4 ligase which were added to the wells, or other factors which would require repeats of this experiment.
Suggestions for ELISA
1. Problem: Absorbances remain unchanged/increases with decreasing protein concentration
Possible cause: Protein concentrates saturates available antibody
Solution 1: Reduce protein concentrations below 1/10000X
Solution 2: Perform a second purification step
Solution 3: Increase antibody concentrations available to bind protein (primary detecting (anti-His) or primary capture (anti-Myc))
2. Problem: Absorbance of positive control is not significantly different from negative controls
Possible cause: steric hinderance of dual tag construct.
Solution 1: add a ALA-6 linker between two tags.
Solution 2: put tags on the opposite side of the protein.
second possible cause: background noise.
solution 1: do more replicates.
TECAN data
Legend
- X axis = Cycles (each cycle is approximately 5 minutes)
- Y axis = Absorbance (relative units, at 600 nm)
- A,F (blue) = GP2578
- B,G (red) = GP2599
- C,H (green) = GP2619
Without arabinose induction
 |
 |
 |
With arabinose induction
 |
 |
 |
TECAN Analysis
Cells did not show differential growth rates with arabinose induced constructs, as expected. No growth was observed for many wells that are near the edges of the TECAN plate; recommend to not use edge wells in future analyses.
4/26/2010
Did ELISA. Left lab around 5 pm; last step performed was incubating 1h after adding varying concentrations of T4 ligase to 24 wells.
Protocol (roughly):
- Dilute anti-Myc antibody (1000X) to 1X in TBST, plate 100 uL into well
- Incubate at RT for 1h
- Do three plate washes (TBST with 0.1% Tween)
- Put blocking buffer (2% BSA, in TBST) 100 uL in wells, incubate 1h at RT
- Do three plate washes
- Put T4 Ligase at various concentrations (diluted in blocking buffer) (see attached spreadsheet) into wells
- Incubate 1h at 37C
- Do three washes
- Put anti-His antibody conjugated to HRP (10000X) 100uL, dilute to 1X in blocking buffer, 1h at 37C
- Do three washes
- Put detection 3,3’,5,5’-Tetramethylbenzidine (TMB) solution 100uL
- Incubate at 37C for 15 min
- Detect with spectrometer at 450 nm
4/22/2010
- Get column
- Pour in lysate + beads
- Collect drips
- Wash with ice cold PBS (20 mL) (N x 0.5 mL)
- Elute with 4-5 1 mL volumes of 300 mM imidazole in PBS
- Put eluant into dialysis column, add ice cold PBS to column, then shake in a speed of 5000mph with 4°C for 30 min, then change to fresh PBS, repeat shaking and change media for several times.
- add ligase, purify and run in gel (Mike did this).
4/21/2010

- General protocol for protein purification:
- Pick starter colony
- Grow starter culture
- Put in flask
- Grow to midlog
- Induce arabinose
- Grow to saturation, have protein
- Transfer to centrifuge tube (100 mL) x 2 - stuff here on is on ice (4C)
- Spin 5 min at 6000 rpm
- Dump supernatant, leave pellet
- 10 mL of resuspension buffer (TBS), resuspend and combine in 50 mL conical flask
- Sonicate (probe) - 1 minute (is it a program?). DO pulses.
- Spin to remove debris
- Transfer supernatant to fresh tube (pour). Will be translucent.
- Add 400 uL of Ni resin
- Agitate at 4C 1h to overnight
- Tomorrow
- Get column
- Pour in lysate + beads
- Collect drips
- Wash with PBS (20 mL) (4 x 5 mL)
- Elute with 4-5 1 mL volumes of 300 mM imidazole in PBS
- Put eluant into dialysis bag
- Let dialyze overnight on ice (4°C)
4/19/2010
- analyze TECAN data, maximize TECAN protocol
- TECAN data showed significant edge effects. Recommended to not use outer rows and columns
- I have no clue how to derive the actual growth rate. Tried some logistic equations (a*b)/(a-(b-a)*Exp(-c*t)) and nonlinear regression but my system keeps blowing up, regression looks bad, etc.
- There are a lot of forms of logistic equations you can use. E.x. K / (1 + exp(a + b*x), the differential form dN/dt = r*N(K-N)/K, etc. If anyone has a copy of Mathematica or SPSS, try to play around with it.
- develop ELISA protocol
4/15/2010
- materials: 20% 100X arabinose, LB media and 96 well plate for TECAN
- dilute arabinose to 1X, 50ul LB in each well. 1ul of saturated cell mixture.
- Plate: http://spreadsheets.google.com/ccc?key=0AvbXytCFRyFCdGJNVXhzNTUtWTgtZFlkYmZGTVQxT2c&hl=en
- by accident, added two uls of different cells to F3
TECAN

Running a TECAN analysis:
The procedure below only works for black 96-well flat bottom plates.
- Turn TECAN power on (wait for light to stop flashing)
- Open XFluor4 Safire II XLS spreadsheet
- Load plate:
- Have 96-well plate with media+cells
- Goto XFluorSafireII menu > movements > out
- Load plate
- Goto XFluorSafireII menu > movements > in
- Load the program to run:
- Select Multi Labeling Kinetic
- Load multi labeling kinetic parameter > "\iGEM 2007\My Documents\Weston\gabe's experimental folder\kinteticmodified_no gfp reading.mps"
- Click "Run"
- Wait for data collection. Operation can only be cancelled when machine is performing measurements.
4/14/2010
Picked 3 colonies from the 3 different constructs. Transformation efficiency is, of course, insane because there was no ligation performed.
 |
 |
 |
Ugh, image markup is confusing. Anyways, refer to the edit page for code to do the above.
4/12/2010
- Trial run for transformation. Obtained vectors.
- jtk2559-jtk2164 K
- jtk2619-jtk2164 K
- jtk2578-jtk2164 K
- Transformation protocol:
- Add:
- plasmid 0.5 uL
- cells (MC1061 pir+) 10 uL
- Add to cells:
- KCM 1.5 uL
- Water 2.5 uL
- Use normal heat shock
- Did @ 42C for 120 sec
- Rescue (rescued 40 min)
- Plate
Wei-Chun Hu 19:36, 31 March 2010 (EDT)
Our team page: Template:SBB10AssayTeam1
Team notes: SBB10AssayTeam1-Notes
Part Synthesis (aka Wet Lab)
Wei-Chun Hu 23:47, 17 March 2010 (EDT)
Chose sbb13-1 and sbb13-2 for sequencing. Put into clotho.
Wei-Chun Hu 00:42, 17 March 2010 (EDT)
Protocol
Awaiting sequence results. Repeat Miniprep if needed. Redo sbb13 (as I predicted), depending on what JCA says.
Wei-Chun Hu 19:27, 15 March 2010 (EDT)
Products
- sbb11-1, sbb11-2, sbb11-3, sbb11-4 - 4 clones picked from sbb11 plate
- sbb13-1, sbb13-2, sbb13-3, sbb13-4 - 4 clones picked from sbb13 plate
- Very (VERY) rough estimation of transformation efficiency:
- sbb11 - (87*17 cells)/(((50 ng))*(1 uL/8.5uL)) = 251 cells/ng insert = 23 cells/ng plasmid
- sbb13 - (42*17 cells)/(((80 ng)*2)*(1 uL/8.5uL)) = 38 cells/ng insert = 17 cells/ng plasmid
- sbb11 transformed nearly 10x as efficiently (gramwise), and approximately the same efficiency molewise (1812/226).
- Choose: sbb11-2, sbb13-4 for sequencing
- Lanes 2-5 look fine (Expected: 1727/684 (EcoRI/XhoI), actual ~1700/650) - chose sbb11-2 (lane 3)
- Lanes 10-13 - (Expected: 2170/1827 (EcoRI/BamHI), actual (sbb13-4): ~2500/1750) - need more resolution to be sure. Chose sbb13-4. If it fails, choose sbb13-1 or sbb13-2.
Wei-Chun Hu 03:07, 15 March 2010 (EDT)
Protocol
Assume white colonies are present. This may or may not be a reasonable assumption.
Miniprep purification of DNA using Macherey-Nagel Nucleospin Kit
MINIPREP (1mL - 5mL) Procedure for Plasmid DNA Purification
- Pellet the cells (for individual cultures)
- Pellet around 1.5 mL to 3 mL saturated culture by spinning full speed, 30 seconds.
- Dump supernatant, repeat to pellet another 1.5 mL (for a total of 3 mL if desired)
- Pellet the cells (for 24-well blocks)
- In 327, weigh the block on the scale, fill a dirty block with water to the same weight
- Use the Allegra centrifuge to spin at 5500rpm for 4 minutes
- Dump supernatant
- You will need to transfer the cells to microcentrifuge tubes after resuspension in A1 in the alkaline lysis section below
- Do an alkaline lysis
- Add 250uL of A1 buffer into each tube. Resuspend the cells using a vortexer.
- Add 250uL of A2 buffer (a base that denatures everything and causes cells to lyse). Gently mix up and down. Solution should become clearer.
- Add 300uL of A3 buffer (an acid of pH ~5 that causes cell junk - including protein and chromosomal DNA - to precipitate, and leaves plasmids and other small molecules in solution). Slowly invert a few times, then shake.
- Spin in centrifuge at top speed for 5 minutes.
- Label columns with an alcohol-resistant lab pen.
- Purify DNA on a column
- Pour liquid into columns, and place the columns into the centrifuge. Spin at full speed for 15 seconds.
- Dump liquid out of the collectors under the columns (the DNA should be stuck to the white resin)
- Wash each column with 500 uL of AW buffer.
- Spin in centrifuge at full speed for approximately 15 seconds, then flick out the liquid again.
- Wash with 750uL of A4 buffer (washes the salts off the resins).
- Spin in centrifuge at full speed for approximately 15 seconds and flick out liquid again.
- Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol.
- Label new tubes and put columns in them.
- Elute them by squirting 50uL of water down the middle of the column (don't let it stick to the sides).
- Spin in centrifuge at top speed for 30 seconds.
- Take out columns and cap the tubes.
- Clean up - note the A1 buffer is stored at 4degC and all the rest at room temperature.
Wei-Chun Hu 14:07, 11 March 2010 (EST)
Products
- 10D - transformed cells from 9D ligation rxn with pBjk2741
- 8ABC - transformed cells from 7ABC ligation rxn with pBjk2741
I don't know how well transformed cells keep under freezing. They are in box C regardless.
Notes
- Strange "spots" noted on Spec+ plates. Could be contamination, but most likely air bubbles.
- 2YT media from last lab was contaminated. Samples should not be affected.
Wei-Chun Hu 18:02, 9 March 2010 (EST)
Protocol
Protocols from here on out should be the same for both procedures, excepting miniprep digest mapping.
Ligation
- Set up the following reaction:
6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest (pBjk2741-Bca1144) 1uL Insert digest (9D or 7ABC) 0.5uL T4 DNA Ligase
- Pound upside down on the bench to mix
- Give it a quick spin to send it back to the bottom of the tube
- Incubate on the benchtop for 30min
- Put on ice and proceed to the transformation
Transformation by heat-shock
Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock.
1. Thaw a 200 uL aliquot of cells on ice 2. Add 50 uL of water to the cells 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two 5. Add 70 uL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min 7. Heat shock for 90 seconds at 42 (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour 10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight
Wei-Chun Hu 19:14, 8 March 2010 (EST)
Products
- 9D - Zymo prep gel of digest 7D1
- 7ABC - Zymo prep gel of digest 5ABC2
- Lane 2: Faint, ~250bp
- Lane 3: Strong, ~1800bp
Wei-Chun Hu 19:59, 7 March 2010 (EST)
Protocol: PCA
- Digest the product (7D1) and desired backbone with Eco/Bam:
- Set up the following reaction (two tubes):
- 8uL of eluted PCR product (backbone [pBjk2741-Bca1144] and PCA product in SEPARATE reactions)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Set up the following reaction (two tubes):
- Incubate 1h
- Load prep. gel
- Zymo g.p.
- cut out bands minimizing extra gel matter.
- put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle).
- heat at 55, shake and/or vortex until the gel has dissolved.
- If the DNA is <300bp add 250uL of isopropanol
- transfer into the Zymo column inside a collection tube (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with 8.5 uL of water into a fresh Eppendorf tube
Protocol: SOE
- Digest the product (5ABC2) and desired backbone with Eco/Bam:
- Set up the following reaction (two tubes):
- 8uL of eluted PCR product (backbone [pBjk2741-Bca1144] and PCA product in SEPARATE reactions)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Set up the following reaction (two tubes):
- Incubate 1h
- Load prep. gel
- Zymo g.p.
- cut out bands minimizing extra gel matter.
- put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle).
- heat at 55, shake and/or vortex until the gel has dissolved.
- transfer into the Zymo column inside a collection tube (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with 8.5 uL of water into a fresh Eppendorf tube
Wei-Chun Hu 13:15, 4 March 2010 (EST)
Overall schedule outline
- Monday
- Digest Eco/Bam (1h)
- Prep. gel
- Zymo gel purification
- Wednesday
- Ligate part and vector digests (30 min)
- Transformation (rescue 1h)
- Plate
- Weekend
- (GSI) Pick colonies
- Next Monday
- Miniprep
- Analytical gel
- Sequence
- Done!
Wei-Chun Hu 17:15, 3 March 2010 (EST)
Products Zymo-ed
Chosen products bolded
- 7D1 - Zymo of JHD (7D)
- 5D1 - Zymo of JHD2 (5D)
- 5ABC2 - Zymo of H2O (4ABC2)
- 5ABC3 - Zymo of DMSO (4ABC3)
Gels
- LADDER
- sbb22 H2O
- sbb22 DMSO
- sbb20 H2O
- sbb20 DMSO
- sbb7 1/2 DMSO
- sbb7 1/2
- JH7D
- JH5D
- JH4ABC2
- JH4ABC3
- AS123
- LADDER
- Band for 7D and 5D look identical. I will choose 7D regardless because it has been purified more.
- Band for 4ABC2 (H2O) and 4ABC3 (DMSO) are nearly identical except for some residual lower bp products in both. H2O looks slightly better than DMSO; however, both are usable.
Wei-Chun Hu 04:16, 2 March 2010 (EST)
A public service announcement
Jim will be gone early on Wed and Fri for personal matters.
"Suggested" Protocol
- Immediately set up 4 analytical gels for all PCRs.
- Select the better out of (4ABC2 [h2o] or 4ABC3 [DMSO]) and (JHD [7D] or JHD2 [5D]).
- Try to get the gels run ASAP. I really don't have much time.
- If at least 2 out of 4 are satisfactory, do a zymo cleanup (if I leave at 3, that's probably as far as I will get.)
Wei-Chun Hu 16:38, 1 March 2010 (EST)
Protocol changes
SOE
Do PCR starting from 3ABC with alternate temperature settings. 2 PCRs done, one with H2O, one with added DMSO. See Template:SBB-Protocols_PCR2.
PCA
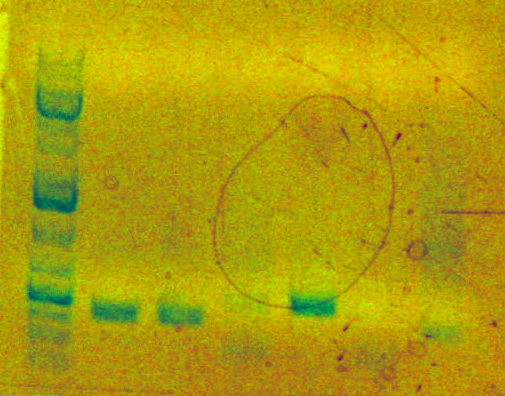
Prep. gel done for 5D. Product is present but in unacceptably low concentration (3rd lane from right):
Zymo gel purification still performed. Eluted with 10 uL ddH2O into 7D.
Both 5D and 7D were used for Re-assembly. Abnormally large quantities of both (5 uL) were used for the PCRs, in place of water:
37 uL32 uL ddH2O- 5 ul 10x expand buffer
- 5 ul 2mM dNTPs
- 1 ul oligo mixture (100uM total, mixture of oligos after combination of 100uM stocks)
1 uL5 uL of purified assembly reaction from previous round- 0.75 ul Expand polymerase
Note that I will need to do another digestion assuming the next prep. gel turns out right (hence, the 5D step is basically useless). It should be obvious why.
Products
- 4ABC2 - new PCR with H2O @ 2K45
- 4ABC3 - new PCR with DMSO @ 2K45
- 5D - digested 4D completed
- 6D,7D - zymo of 5D. Elute using 10 uL of ddH2O.
- (both 5D and 7D were used for PCR - 5 uL of each rxn).
- JHD (PCR) = uses 5 uL of 7D
- JHD2 (PCR) = uses 5 uL of 5D
Wei-Chun Hu 17:13, 28 February 2010 (EST)
Protocol
- The big picture: Proceed with PCA. Do not proceed with SOE.
Protocol: PCA
- Digest the product and desired backbone with Eco/Bam:
- Set up the following reaction (two tubes):
- 8uL of eluted PCR product (already in tube 5D, do NOT put more)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Set up the following reaction (two tubes):
- Incubate, wait for 1hr
- Run a prep. gel
- Cut out bands, zymo gel purification:
- put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle).
- heat at 55, shake and/or vortex until the gel has dissolved.
- Add 250 uL of isopropanol.
- transfer into the Zymo column inside a collection tube (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- discard collection tube
Wei-Chun Hu 18:18, 24 February 2010 (EST)
Protocol changes
- Change for protocol (replace with small fragment zymo cleanup) for PCA
- Did not get to Digest step (advised to hold off due to running time, and given possible failure of 4ABC...)
Products
- 4D - zymo-ed PCA (band looks fine, but faint)
- 5D - incomplete digest reaction - contains 8 uL of 4D
- 5ABC - zymo-ed SOEing PCR (problem: see below)
- 6ABC - incomplete digest reaction - contains 8 uL of 5ABC
Potential problems
- Band for 4ABC analytical gel is WAY off (5th from the left band is a bit over 500, product should be around 1800). Jeni also had the same problem with SOEing (band on far right). Recommended change SOEing protocol (SLIC, Venter?)
Wei-Chun Hu 15:31, 23 February 2010 (EST)
Protocol: PCA
- Optional: Run an analytical gel - 3 uL PCR, 7 uL loading buffer
- This gel is probably not necessary because we will do a gel in the digest step
- Band should be around 250 bp (below the 1st bright one)
- Clean up the amplification reactions with a zymo column
- Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction.
- Transfer into the Zymo column (small clear guys)
- Add 500uL of Ethanol and pipette up and down to mix
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube
- Digest the product and desired backbone with Eco/Bam:
- Set up the following reaction (two tubes):
- 8uL of eluted PCR product (backbone [pBjk2741-Bca1144] and PCA product in SEPARATE reactions)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Incubate at 37 degrees on the thermocycler for 1hr
- Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an acceptable stopping point
- Only proceed below if ample time
- Set up a Zymo column
- The DNA is shorter than 300bp: add 250uL of isopropanol and mix prior to loading it on the column
- Set up the following reaction (two tubes):
Protocol: SOE
- Optional: Run an analytical gel - 3 uL PCR, 7 uL loading buffer
- Expect bright band around 1800 bp (just above the middle bright one)
- Clean up the amplification reactions with a zymo column
- Add 180 uL of Zymo ADB buffer (brown bottle) to the reaction.
- Transfer into the Zymo column (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube
- Digest the product and desired backbone with Eco/Bam:
- Set up the following reaction (we already have the plasmid digest):
- 8uL of eluted PCR product (PCR product)
- 1uL of NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
- Incubate at 37 degrees on the thermocycler for 1hr
- Run an agarose gel, and melt with 600uL ADB buffer at 55 degrees. ****NOTE: If you are running short of time, this is an acceptable stopping point
- Set up the following reaction (we already have the plasmid digest):
Wei-Chun Hu 17:31, 22 February 2010 (EST)
Results
Gel for PCR
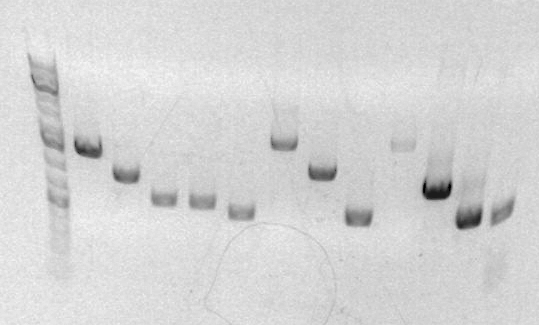
Last three columns are A,B,C. Lengths appear to be correct (~900,~500,~450). Distortion of gel makes it impossible to accurately compare the last two.
Protocol changes
8uL6uL of PCR product premixed with1uL4uL of loading buffer- Used Jeni's mastermix for 4ABC PCR because frankly everyone was running out of time
Products
- PCA
- 2D - zymo cleanup of 1D
- 3D - PCR amplification of 2D (put in PCA box)
- SOE
- JHA,JHB,JHC - preparative gels of 1A,1B,1C (6 uL PCR product / 4 uL 1X loading buffer)
- 2ABC - purifying gel fragments 1A,1B,1C
- 3ABC - purified 50uL DNA from gel fragments
- 4ABC - PCR reaction using 0.5 uL of 3ABC (put in 2K55 box)
Wei-Chun Hu 18:21, 21 February 2010 (EST)
Protocol: SOE
(This assumes the PCR has completed successfully.)
- For each PCR, load 6uL of PCR product premixed with 4uL of loading buffer in a single well of a 1% agarose gel
- Cut out the bands, put them into a single 1.5mL microcentrifuge tube (put ALL bands in one tube)
- Add 650uL of ADB Buffer
Zymo gel purification
- All spins until the drying step are 15 second full speed spins.
cut out bands minimizing extra gel matter.put in ependorf tube and add 600uL of Zymo ADB buffer (brown bottle).- heat at 55, shake and/or vortex until the gel has dissolved.
- (do NOT put isopropanol.)
- transfer into the Zymo column inside a collection tube (small clear guys)
- spin through, discard waste.
- Add 200 uL of PE buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- discard collection tube
- Elute the DNA in 50uL of water in a LABELED eppendorf tube
- Dilute 10 uM stocks of OJIMH011 and OJIMH014.
- Set up your second round of PCR as a normal 33uL reaction using the eluted mixture of fragments as template:
24uL ddH2O 3.3uL 10x Expand Buffer "2" 3.3uL dNTPs (2mM in each) 1uL OJIMH011, 10uM (dilute 100 uM stock) 1uL OJIMH014, 10uM (dilute 100 uM stock) 0.5uL Expand polymerase "1" 0.5uL Template DNA
Protocol: PCA
Small-Frag Zymo Cleanup
The following procedure removes the polymerase, dNTPs, buffer, and most of the oligonucleotides from a PCR reaction for fragments smaller than 300bp. It also will remove the buffer and restriction enzymes from a restriction digest reaction.
- Add 100 uL of Zymo ADB buffer (brown bottle) to the reaction.
- Transfer into the Zymo column (small clear guys)
- Add 500uL of Ethanol and pipette up and down to mix
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer (which is basically 70% ethanol)
- spin through, discard waste.
- Add 200 uL of PE or Zymo Wash buffer
- spin through, discard waste.
- spin for 90 seconds, full speed to dry.
- elute with water into a fresh Eppendorf tube
Amplification
Now, you need to do an amplification of the correct full-length chunks. Clean up the assembly reaction with a zymo column; don't bother running it on a gel - it'll be a smeary mess and won't really help you. Save the purified product in case this step fails! For the amplification reaction, do a normal phusion program with 1 ul of the cleaned up assembly reaction as template, and using the outermost oligos for the chunk. That is:
Recipe
- 1 ul each outer oligo [sbb11 F/R] (10 uM)
- .5 ul phusion
- 10 ul 5x phusion buffer
- 5 ul 2mM dNTPs
- 32.5 ul H2O
Program
- 2 min initial denature at 94oC
- 30 sec denature at 94oC
- 30 sec anneal at 60oC [This should be high, as your outer oligos now have a huge overlap with the correct product]
- 30 sec extension at 68oC
- repeat 2-4 30 times total
Wei-Chun Hu 17:33, 17 February 2010 (EST)
Protocol
Resuspend lypophilized DNA. Oligo stock is 100 uM, dilute to 10 uM.
Appropriate components (see Media:2-17-10-cloning-PCR.pdf) were added to three Eppendorf tubes. Transfer to PCR tubes at end. DNA polymerase was added to the mastermix.
Oligos for PCA were provided as mixture (100 uM, M), forward (10 uM, F), and reverse (10 uM, R). The Template:SBB-PCA protocol was used.
Products
1A: OJIMH011,OJIMH013,Bca1623 6-9
1B: OJIMH012,CA1674,Bca1559 7-7
1C: CA1675,OJIMH014,Bca1559 7-1
1D: Oligo mixture (M SBB11)
Wei-Chun Hu 18:10, 16 February 2010 (EST)
Run this protocol tomorrow: Media:2-17-10-cloning-PCR.pdf
Wei-Chun Hu 14:24, 9 February 2010 (EST)
The following protocols are relevant for me:
Cloning by PCR
SOEing PCR
PCA Gene Synthesis
Regular Zymo Cleanup
Zymo Gel Purification
EcoRI/BamHI Digest of PCR Products
Ligation of EcoRI/BamHI digests
Transformation by heat-shock
Picking of colonies
Miniprep purification of DNA
Computational Part Design
Wei-Chun Hu 01:45, 4 February 2010 (EST)
All construction files (now transcluded)
1) sbb11: sleeping beauty 5'TR Pool OJIMH001 through OJIMH010, assemble by PCA PCR OJIMH001/OJIMH010 on PCA reaction (257 bp, EcoRI/BamHI) Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L) Product is pBjk2741-sbb11 ------------ OJIMH001 PCA assembly of sbb11 CCATAGAATTCATGAGATCTAGTTGAAGTCGGAAGTTTACATACACTTAAG OJIMH002 PCA assembly of sbb11 GAAAAACGAGTTTTAATGACTCCAACTTAAGTGTATGTAAACTTCCGACTTC OJIMH003 PCA assembly of sbb11 AAGTTGGAGTCATTAAAACTCGTTTTTCAACTACACCACAAATTTCTTGTTAAC OJIMH004 PCA assembly of sbb11 CTGACTTGCCAAAACTATTGTTTGTTAACAAGAAATTTGTGGTGTAGTTGAAA OJIMH005 PCA assembly of sbb11 TTAACAAACAATAGTTTTGGCAAGTCAGTTAGGACATCTACTTTGTGC OJIMH006 PCA assembly of sbb11 ACAATTGTTGGAAAAATGACTTGTGTCATGCACAAAGTAGATGTCCTAACTG OJIMH007 PCA assembly of sbb11 GACACAAGTCATTTTTCCAACAATTGTTTACAGACAGATTATTTCACTTATAATTC OJIMH008 PCA assembly of sbb11 CCACTGGAATTGTGATACAGTGAATTATAAGTGAAATAATCTGTCTGTAAAC OJIMH009 PCA assembly of sbb11 TAATTCACTGTATCACAATTCCAGTGGGTCAGAAGTTTACATACACTAAG OJIMH010 PCA assembly of sbb11 CTGATGGATCCTTAGTGTATGTAAACTTCTGACCC
JCA Notes
- Correct
2) sbb13: PhiC31 Integrase
PCR OJIMH011/OJIMH012 on pBca9523-Bca1623 (949 bp, gp = A)
PCR OJIMH013/OJIMH014 on pBca9523-Bca1659 (938 bp, gp = B)
-------------
PCR PJIMH011/OJIMH014 on A+B (1843 bp, EcoRI/BamHI)
Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L)
Product is pBjk2741-sbb13 {<phiC31>}
-------------
OJIMH011 Forward PCR of Part 1 of PhiC31 ccataGAATTCatgAGATCTGACACGTACGCGGGTGCTTA
OJIMH012 SOEing of Part 1 (R) of PhiC31 AAGAAGCCGGACGGCACGCCGACCacgaagattgagggttaccg
OJIMH013 SOEing of Part 2 (F) of PhiC31 cggtaaccctcaatcttcgtGGTCGGCGTGCCGTCCGGCTTCTT
OJIMH014 Reverse PCR of Part 2 of PhiC31 atcagGGATCCCGCCGCTACGTCTTCCGT
JCA Notes
- Oligos 12 and 13 are switched
- Otherwise correct
sbb13: PhiC31 Integrase PCR OJIMH011/OJIMH013 on Bca1623 6-9 (949bp, gp=A) PCR OJIMH012/CA1674 on Bca1559 7-7 (514bp, gp=B) PCR CA1675/OJIMH014 on Bca1559 7-1 (444bp, gp = C) ------------- PCR PJIMH011/OJIMH014 on A+B+C (1843 bp, EcoRI/BamHI) Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L) Product is pBjk2741-sbb13 {<phiC31>} ------------- OJIMH011 Forward PCR of Part 1 of PhiC31 ccataGAATTCatgAGATCTGACACGTACGCGGGTGCTTA OJIMH012 SOEing of Part 1 (R) of PhiC31 AAGAAGCCGGACGGCACGCCGACCacgaagattgagggttaccg OJIMH013 SOEing of Part 2 (F) of PhiC31 cggtaaccctcaatcttcgtGGTCGGCGTGCCGTCCGGCTTCTT CA1674 GGGCGTCGGCGCGCTCCGCAACAAGGTTCGCCCGTTCGCCGCTCTTCTCAG PCA assembly of phiCthreeprime (Bca1659) CA1675 TGCGGAGCGCGCCGACGCCCTGAACGCCCTTGAAGAGCTGTACGAAGACCG PCA assembly of phiCthreeprime (Bca1659) OJIMH014 Reverse PCR of Part 2 of PhiC31 atcagGGATCCCGCCGCTACGTCTTCCGT
Wei-Chun Hu 20:45, 3 February 2010 (EST)
Construction file for sbb13 (this one is especially tricky, so please check)
2) sbb13: PhiC31 Integrase
PCR OJIMH011/OJIMH012 on pBca9523-Bca1623 (949 bp, gp = A)
PCR OJIMH013/OJIMH014 on pBca9523-Bca1659 (938 bp, gp = B)
-------------
PCR PJIMH011/OJIM014 on A+B (1843 bp, EcoRI/BamHI)
Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L)
Product is pBjk2741-sbb13 {<phiC31>}
-------------
OJIMH011 Forward PCR of Part 1 of PhiC31 ccataGAATTCatgAGATCTGACACGTACGCGGGTGCTTA
OJIMH012 SOEing of Part 1 (R) of PhiC31 AAGAAGCCGGACGGCACGCCGACCacgaagattgagggttaccg
OJIMH013 SOEing of Part 2 (F) of PhiC31 cggtaaccctcaatcttcgtGGTCGGCGTGCCGTCCGGCTTCTT
OJIMH014 Reverse PCR of Part 2 of PhiC31 atcagGGATCCCGCCGCTACGTCTTCCGT
Wei-Chun Hu 20:03, 3 February 2010 (EST)
Construction file for sbb11 (check for errors)
1) sbb11: sleeping beauty 5'TR Pool OJIMH001 through OJIMH010, assemble by PCA PCR OJIMH001/OJIMH010 on PCA reaction (257 bp, EcoRI/BamHI) Sub in pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L) Product is pBjk2741-sbb11 ------------ OJIMH001 PCA assembly of sbb11 CCATAGAATTCATGAGATCTAGTTGAAGTCGGAAGTTTACATACACTTAAG OJIMH002 PCA assembly of sbb11 GAAAAACGAGTTTTAATGACTCCAACTTAAGTGTATGTAAACTTCCGACTTC OJIMH003 PCA assembly of sbb11 AAGTTGGAGTCATTAAAACTCGTTTTTCAACTACACCACAAATTTCTTGTTAAC OJIMH004 PCA assembly of sbb11 CTGACTTGCCAAAACTATTGTTTGTTAACAAGAAATTTGTGGTGTAGTTGAAA OJIMH005 PCA assembly of sbb11 TTAACAAACAATAGTTTTGGCAAGTCAGTTAGGACATCTACTTTGTGC OJIMH006 PCA assembly of sbb11 ACAATTGTTGGAAAAATGACTTGTGTCATGCACAAAGTAGATGTCCTAACTG OJIMH007 PCA assembly of sbb11 GACACAAGTCATTTTTCCAACAATTGTTTACAGACAGATTATTTCACTTATAATTC OJIMH008 PCA assembly of sbb11 CCACTGGAATTGTGATACAGTGAATTATAAGTGAAATAATCTGTCTGTAAAC OJIMH009 PCA assembly of sbb11 TAATTCACTGTATCACAATTCCAGTGGGTCAGAAGTTTACATACACTAAG OJIMH010 PCA assembly of sbb11 CTGATGGATCCTTAGTGTATGTAAACTTCTGACCC
Wei-Chun Hu 19:45, 3 February 2010 (EST)
A test entry.