SBB10Ntbk-RaffiHagopian: Difference between revisions
No edit summary |
|||
| (20 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
See http://openwetware.org/wiki/Team_2_Notes for transposase assay team notes. | |||
Construction files for the parts I'm making: | Construction files for the parts I'm making: | ||
| Line 24: | Line 26: | ||
</pre> | </pre> | ||
=='''3-17-2010'''== | |||
I got back my sequencing data today. The read for my sbb09 part seems fine but had some ambiguity near the BamHI site. The professor suggested resequencing this sample. The read for my sbb33 part had a missense mutation in it so we have to sequence another clone. | |||
I was also able to plate colonies from my 3-8-2010 sbb33 spec plate onto some gentamicin plates. Hopefully some of my clones will survive this time (recall that no colonies were seen on the 3-8-2010 gentamicin plate). | |||
'''UPDATE:''' | |||
Although some colonies did appear to grow on the gentamicin plate it is possible that they were contaminants as they looked a little like yeast. The professor mentioned that he can find a GenR part from somewhere else. Hence I stopped worrying about sbb33. | |||
The sbb09 part seems fine. | |||
=='''3-15-2010'''== | |||
I submitted samples 3 & 4 for part sbb09 and sample 2 & 3 for part sbb33 for sequencing. | |||
=='''3-12-2010'''== | |||
I set up analytical digest reactions for all of my miniprep plasmid samples (four for part sbb33 and four for sbb09). | |||
Part sbb33 is 833bp so I incubated it with EcoRI/BamHI. | |||
Part sbb09 is a short EIPCR part so it was EcoRI/XhoI. | |||
The gel indicated that samples 1 to 4 of the sbb09 mapped as expected. | |||
Sample 2 to 4 of part sbb33 also mapped as expected. | |||
'''Mapping Gel:''' | |||
[[Image: RH-3-12-10.jpg]] | |||
#Ladder | |||
#RH-9-MAP-1 | |||
#RH-9-MAP-2 | |||
#RH-9-MAP-3 | |||
#RH-9-MAP-4 | |||
#RH-33-MAP-1 | |||
#RH-33-MAP-2 | |||
#RH-33-MAP-3 | |||
#RH-33-MAP-4 | |||
#Ladder | |||
I am done with the construction of my basic parts. I will submit sample 3 and 4 for part sbb09 for sequencing and sample 2 and 3 for part sbb33 for sequencing. | |||
=='''3-10-2010'''== | =='''3-10-2010'''== | ||
I got back my three plates. The Sbb33 with gentamicin and spec plate produced no colonies. | I got back my three plates. The Sbb33 with gentamicin and spec plate produced no colonies. | ||
The sbb33 spec plate and sbb09 spec plate did produce colonies (4 colonies were picked from each plate). *Contamination NOTE* The LB media I used on Monday was apparently contaminated. Small white contaminants can be seen on my spec plates. | The sbb33 spec plate and sbb09 spec plate did produce colonies (4 colonies were picked from each plate). No red colonies appeared on my sbb33 spec plate. The ratio of red to white colonies on my sbb09 spec plate was around ~1/25. *Contamination NOTE* The LB media I used on Monday was apparently contaminated. Small white contaminants can be seen on my spec plates. | ||
I completed the miniprep purification protocol for my 8 colony samples (4 for part sbb09 and 4 for sbb33). | I completed the miniprep purification protocol for my 8 colony samples (4 for part sbb09 and 4 for sbb33). | ||
The eight samples were stored in Box B. *Miniprep Note* I used 1.5 mL of cells instead of the suggested 3mL of cells initially because I wanted to save some cells for backup purposes. | The eight samples were stored in Box B. *Miniprep Note* I used 1.5 mL of cells instead of the suggested 3mL of cells initially because I wanted to save some cells for backup purposes. | ||
=='''3-8-2010'''== | =='''3-8-2010'''== | ||
| Line 41: | Line 83: | ||
Part sbb33 was plated twice, once on spec and also on another plate containing both spec and gentamicin. | Part sbb33 was plated twice, once on spec and also on another plate containing both spec and gentamicin. | ||
=='''3-3-2010'''== | =='''3-3-2010'''== | ||
| Line 48: | Line 89: | ||
Once the digest had incubated for an hour, I set up a preparative gel sample to purify the digested DNA. I was able to complete the entire zymo gel purification protocol by the end of class. The clean DNA sample was placed in a tube labeled "RH-33-Clean-DIG". | Once the digest had incubated for an hour, I set up a preparative gel sample to purify the digested DNA. I was able to complete the entire zymo gel purification protocol by the end of class. The clean DNA sample was placed in a tube labeled "RH-33-Clean-DIG". | ||
'''Gel for sbb33 EcoRI/BamHI digest:''' | |||
[[Image:MN030310Agel.png]]<br> | |||
#Ladder | |||
#'''RH33-DIG''' | |||
#sbb01 | |||
#sbb02 | |||
#Ladder | |||
=='''3-1-2010'''== | =='''3-1-2010'''== | ||
I made analytical gel samples for the SOEing reactions for sbb33 I set up last session. The gel results showed the presence of a ~833bp product so the PCR reaction was a success. | I made analytical gel samples for the SOEing reactions for sbb33 I set up last session. The gel results showed the presence of a ~833bp product so the PCR reaction was a success. | ||
'''Gel for SOEing Sbb33 A+B product:''' | |||
[[Image:MN030110.jpg]]<br> | |||
#Ladder | |||
#'''RH33-A+B-2K45''' | |||
#'''RH33-A+B-2K45 w/ DMSO''' | |||
#sbb02 | |||
#sbb01 | |||
#sbb03 | |||
#JH50 | |||
#PiggyBacA | |||
#Ladder | |||
I performed zymo cleanups on the 2k45 and 2k45 DMSO versions of A+B for part sbb33. I tried to rush through the cleanups and as a result I messed up the 2k45 A+B product's cleanup. Fortunately, I still had the 2k45 DMSO A+B PCR product and did a zymo cleanup on it instead. | I performed zymo cleanups on the 2k45 and 2k45 DMSO versions of A+B for part sbb33. I tried to rush through the cleanups and as a result I messed up the 2k45 A+B product's cleanup. Fortunately, I still had the 2k45 DMSO A+B PCR product and did a zymo cleanup on it instead. | ||
| Line 67: | Line 131: | ||
In order to debug the lack of band for sbb33-A from last lab session, I setup preparative gel samples for sbb33A 2k55, 2k45, 2k45 DSMO. No band appeared for 2k55. It appears that the annealing temperature was the reason no band showed up in the previous gel for this PCR product. Bands appeared for sbb33A when the 2k45 and 2k45 DMSO PCR protocols were used. I cut out the sbb33-A product from the gel, mixed it with the sbb33-B cut out, and performed a gel zymo purification on this mixture. I stored the result in a tube labeled "RH-clean-33-A & B-Temp". | In order to debug the lack of band for sbb33-A from last lab session, I setup preparative gel samples for sbb33A 2k55, 2k45, 2k45 DSMO. No band appeared for 2k55. It appears that the annealing temperature was the reason no band showed up in the previous gel for this PCR product. Bands appeared for sbb33A when the 2k45 and 2k45 DMSO PCR protocols were used. I cut out the sbb33-A product from the gel, mixed it with the sbb33-B cut out, and performed a gel zymo purification on this mixture. I stored the result in a tube labeled "RH-clean-33-A & B-Temp". | ||
'''Preparative Gel for sbb33-A repeated PCR:''' | |||
[[Image:JH022410gelprep.jpg]]<br> | |||
#Ladder | |||
#'''RHA 2K55''' | |||
#'''RHA 2K45''' | |||
#'''RHA 2K45 DMSO''' | |||
#sbb10A | |||
#sbb10B | |||
#FK44 RT | |||
#FK2/4 DMSO | |||
I took the zymo cleaned sbb9 PCR product and did a EIPCR BglII digest on it. I began the gel purification of this digest. I stored the melted gel sample in a tube with 600uL of ADB buffer. | I took the zymo cleaned sbb9 PCR product and did a EIPCR BglII digest on it. I began the gel purification of this digest. I stored the melted gel sample in a tube with 600uL of ADB buffer. | ||
[[ | |||
'''Sbb09 digest gel purification:''' | |||
[[Image:AS22410.png]]<br> | |||
#Ladder | |||
#sbb23 | |||
#JHL(JG?) | |||
#'''RH9 Dig''' | |||
#sbb31 digest | |||
#KRM 27 | |||
#sbb34 | |||
#sbb25 | |||
#Ladder | |||
=='''2-22-2010'''== | =='''2-22-2010'''== | ||
| Line 79: | Line 167: | ||
The analytical gel for RH-PCR-9 produced a band at the expected size (2230bp). | The analytical gel for RH-PCR-9 produced a band at the expected size (2230bp). | ||
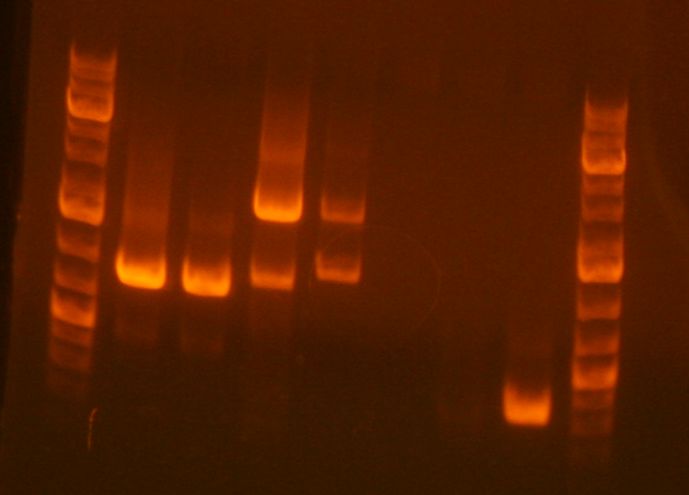
'''Analytical gel for sbb09:''' | |||
[[Image:Zgel_JHL_2_22_10.png]]<br> | |||
Lanes | |||
#ladder | |||
#'''RH9''' | |||
#AS01 | |||
#AS02 | |||
#AS03 | |||
#FK001, 003, PT | |||
#FK004, 002, PT | |||
#sbb07 1/3 | |||
#sbb07 2/4 | |||
#sbb2 A | |||
#sbb2 B | |||
#sbb3 C | |||
#ladder | |||
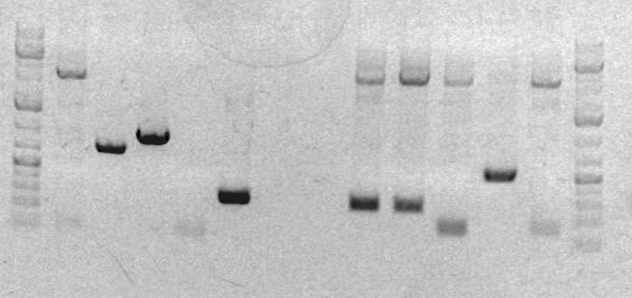
On the preparative gel, RH-PCR-33B produced a band at the expected size (284bp). This band was sliced out of the gel and stored in box "C" in a tube labeled RH-GP-B. RH-PCR-33A failed to produce a band. To debug this issue, I set up three PCR reactions for the sbb33A product (2k55, 2k45, 2k45+DMSO). | On the preparative gel, RH-PCR-33B produced a band at the expected size (284bp). This band was sliced out of the gel and stored in box "C" in a tube labeled RH-GP-B. RH-PCR-33A failed to produce a band. To debug this issue, I set up three PCR reactions for the sbb33A product (2k55, 2k45, 2k45+DMSO). | ||
'''Preparative gel for sbb33-A and sbb33-B:''' | |||
[[Image:XYL022210gel1.jpg]]<br> | |||
#ladder | |||
#sbb04A | |||
#sbb04B | |||
#sbb04C | |||
#'''RH33A''' | |||
#'''RH33B''' | |||
#blank | |||
#blank | |||
#CD1 | |||
#CD2 | |||
#CD3 | |||
#sbb19A | |||
#sbb19B | |||
#ladder | |||
=='''2-17-2010'''== | =='''2-17-2010'''== | ||
Latest revision as of 21:21, 2 May 2010
See http://openwetware.org/wiki/Team_2_Notes for transposase assay team notes.
Construction files for the parts I'm making:
EIPCR rh01F/tn0002R on pBjk2741-Bca1144 (2230 bp, BglII)
Product is pBjk2741-sbb09 {Tn5 3'TR}
----
rh01F EIPCR construction of Tn5 3'TR basic part ccataAGATCTggttgagatgtgtataagagacagtcgacGGATCCtaactcgctcctcag
tn0002R Reverse BglII oligo for Tn5 EIPCR CCAATAGATCTcatgaattcCACTTCAG
Construction of Gentamicin resistance gene (sbb33) BglBrick Part
PCR rh03F/rh05R on pLoxGen4 (576 bp, gp = A)
PCR rh04F/rh06R on pLoxGen4 (284 bp, gp = B)
----
PCR rh03F/rh06R on A+B (bp 833, EcoRI/BamHI)
Sub into pBjk2741-Bca1144 (EcoRI/BamHI, 2170+910, L)
Product is pBjk2741-sbb33 {Promoter.rbs.GenR}
----
rh03F Forward EcoRI for BglBricking GenR ctctgGAATTCatgAGATCTctagcgcgtcgacataagcc
rh04F Removing the BglII site from GenR cgcgtagtgagatAtatatctatgatc
rh05R Removing the BglII site from GenR gatcatagatataTatctcactacgcg
rh06R Reverse BamHI for BglBricking GenR gcaaaGGATCCctagcgcgtcggccgggaag
3-17-2010
I got back my sequencing data today. The read for my sbb09 part seems fine but had some ambiguity near the BamHI site. The professor suggested resequencing this sample. The read for my sbb33 part had a missense mutation in it so we have to sequence another clone.
I was also able to plate colonies from my 3-8-2010 sbb33 spec plate onto some gentamicin plates. Hopefully some of my clones will survive this time (recall that no colonies were seen on the 3-8-2010 gentamicin plate).
UPDATE: Although some colonies did appear to grow on the gentamicin plate it is possible that they were contaminants as they looked a little like yeast. The professor mentioned that he can find a GenR part from somewhere else. Hence I stopped worrying about sbb33.
The sbb09 part seems fine.
3-15-2010
I submitted samples 3 & 4 for part sbb09 and sample 2 & 3 for part sbb33 for sequencing.
3-12-2010
I set up analytical digest reactions for all of my miniprep plasmid samples (four for part sbb33 and four for sbb09). Part sbb33 is 833bp so I incubated it with EcoRI/BamHI. Part sbb09 is a short EIPCR part so it was EcoRI/XhoI.
The gel indicated that samples 1 to 4 of the sbb09 mapped as expected. Sample 2 to 4 of part sbb33 also mapped as expected.
Mapping Gel:
- Ladder
- RH-9-MAP-1
- RH-9-MAP-2
- RH-9-MAP-3
- RH-9-MAP-4
- RH-33-MAP-1
- RH-33-MAP-2
- RH-33-MAP-3
- RH-33-MAP-4
- Ladder
I am done with the construction of my basic parts. I will submit sample 3 and 4 for part sbb09 for sequencing and sample 2 and 3 for part sbb33 for sequencing.
3-10-2010
I got back my three plates. The Sbb33 with gentamicin and spec plate produced no colonies. The sbb33 spec plate and sbb09 spec plate did produce colonies (4 colonies were picked from each plate). No red colonies appeared on my sbb33 spec plate. The ratio of red to white colonies on my sbb09 spec plate was around ~1/25. *Contamination NOTE* The LB media I used on Monday was apparently contaminated. Small white contaminants can be seen on my spec plates.
I completed the miniprep purification protocol for my 8 colony samples (4 for part sbb09 and 4 for sbb33). The eight samples were stored in Box B. *Miniprep Note* I used 1.5 mL of cells instead of the suggested 3mL of cells initially because I wanted to save some cells for backup purposes.
3-8-2010
I performed ligation reactions for both of my basic parts today. I was also able to carry out the heat-shock transformation protocol for both parts.
Part sbb09 was plated on spec. Part sbb33 was plated twice, once on spec and also on another plate containing both spec and gentamicin.
3-3-2010
I performed an EcoRI/BamHI digest on my sbb33 A+B PCR product. While this reaction was incubating, I volunteered to be a gel loader for the day.
Once the digest had incubated for an hour, I set up a preparative gel sample to purify the digested DNA. I was able to complete the entire zymo gel purification protocol by the end of class. The clean DNA sample was placed in a tube labeled "RH-33-Clean-DIG".
Gel for sbb33 EcoRI/BamHI digest:
- Ladder
- RH33-DIG
- sbb01
- sbb02
- Ladder
3-1-2010
I made analytical gel samples for the SOEing reactions for sbb33 I set up last session. The gel results showed the presence of a ~833bp product so the PCR reaction was a success.
Gel for SOEing Sbb33 A+B product:
- Ladder
- RH33-A+B-2K45
- RH33-A+B-2K45 w/ DMSO
- sbb02
- sbb01
- sbb03
- JH50
- PiggyBacA
- Ladder
I performed zymo cleanups on the 2k45 and 2k45 DMSO versions of A+B for part sbb33. I tried to rush through the cleanups and as a result I messed up the 2k45 A+B product's cleanup. Fortunately, I still had the 2k45 DMSO A+B PCR product and did a zymo cleanup on it instead.
My remaining tasks {sbb33:(digest+gp) and sbb09:(ligate+transform)} were both very time intensive hence I decided it would be best to leave them until another lab session.
2-26-2010
Additional lab time was provided today (Friday). I finished the zymo gel purification for the sbb09 part digest.
I also setup my final SOEing PCR reaction using the RH-33-A,B PCR products as template DNA.
2-24-2010
In order to debug the lack of band for sbb33-A from last lab session, I setup preparative gel samples for sbb33A 2k55, 2k45, 2k45 DSMO. No band appeared for 2k55. It appears that the annealing temperature was the reason no band showed up in the previous gel for this PCR product. Bands appeared for sbb33A when the 2k45 and 2k45 DMSO PCR protocols were used. I cut out the sbb33-A product from the gel, mixed it with the sbb33-B cut out, and performed a gel zymo purification on this mixture. I stored the result in a tube labeled "RH-clean-33-A & B-Temp".
Preparative Gel for sbb33-A repeated PCR:
- Ladder
- RHA 2K55
- RHA 2K45
- RHA 2K45 DMSO
- sbb10A
- sbb10B
- FK44 RT
- FK2/4 DMSO
I took the zymo cleaned sbb9 PCR product and did a EIPCR BglII digest on it. I began the gel purification of this digest. I stored the melted gel sample in a tube with 600uL of ADB buffer.
Sbb09 digest gel purification:
- Ladder
- sbb23
- JHL(JG?)
- RH9 Dig
- sbb31 digest
- KRM 27
- sbb34
- sbb25
- Ladder
2-22-2010
We received our PCR products today. I prepared two preparative gel samples for RH-PCR-33-A and B. I also prepared an analytical gel sample for RH-PCR-9. I performed a Regular Zymo cleanup on RH-PCR-9. I stored the clean DNA from this zymo procedure into a tube labeled "RH-9-Clean-PCR" (33uL). The "RH-9-Clean-PCR" tube was placed in "Box 140L C".
The analytical gel for RH-PCR-9 produced a band at the expected size (2230bp).
Analytical gel for sbb09:
Lanes
- ladder
- RH9
- AS01
- AS02
- AS03
- FK001, 003, PT
- FK004, 002, PT
- sbb07 1/3
- sbb07 2/4
- sbb2 A
- sbb2 B
- sbb3 C
- ladder
On the preparative gel, RH-PCR-33B produced a band at the expected size (284bp). This band was sliced out of the gel and stored in box "C" in a tube labeled RH-GP-B. RH-PCR-33A failed to produce a band. To debug this issue, I set up three PCR reactions for the sbb33A product (2k55, 2k45, 2k45+DMSO).
Preparative gel for sbb33-A and sbb33-B:
- ladder
- sbb04A
- sbb04B
- sbb04C
- RH33A
- RH33B
- blank
- blank
- CD1
- CD2
- CD3
- sbb19A
- sbb19B
- ladder
2-17-2010
Today was the first day of wet lab work. I made 100 uM stock solutions for oligos: rh01F,rh03F,rh04F,rh05R,rh06R. I obtained oligo tn0002R from a lab mate. I followed the Cloning by PCR protocol to setup the EIPCR mixture for part sbb09. I followed this same protocol another two times to set up the initial two SOEing PCR mixtures for part sbb33.
At the end of the day I had three tubes ready for PCR:
PCR Tube Label: RH-PCR-9 Description: EIPCR mixture for constructing part sbb09. Program: 4K55 PCR program (it's 2200bp long). PCR Tube Label: RH-PCR-33-A Description: The PCR mixture for obtaining SOEing product A for part sbb33. Program: 2K55 PCR program (it's under 2000bp long). PCR Tube Label: RH-PCR-33-B Description: The PCR mixture for obtaining SOEing product B for part sbb33. Program: 2K55 PCR program (it's under 2000bp long).