SBB10Ntbk-Paulina Tran: Difference between revisions
Paulina Tran (talk | contribs) |
Paulina Tran (talk | contribs) |
||
| Line 100: | Line 100: | ||
==14:30, 3 March 2010 == | ==14:30, 3 March 2010 == | ||
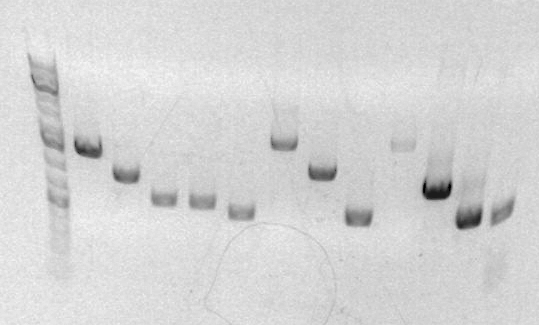
Today, I miniprepped TN1/2R. After finishing, I continued to prep FK 1/3 2/4 for analytical gel. | |||
[[Image:Ak3-3-10.png]] | [[Image:Ak3-3-10.png]] | ||
sbb21 (FK 1/3 2/4) product is expected to have a band at ~600bp. The product is in lane 4 and looks approximately at 600bp. | The sbb21 (FK 1/3 2/4) product is expected to have a band at ~600bp. The product is in lane 4 and looks approximately at 600bp. Therefore the product is good. I will continue with zymo clean up and proceed with anEcoI/BamHI digestion next week. | ||
==14:00, 8 March 2010 == | ==14:00, 8 March 2010 == | ||
Revision as of 03:06, 29 April 2010
Paulina Tran 02:00, 9 February 2010
Construction file for sbb21
Construction of FokI+ cleavage domain basic part sbb21
PCR fk0001/fk0003 on K243000 (505 bp, gp = A)
PCR fk0004/fk0002 on K243000 (141 bp, gp = B)
---------------------------------
PCR fk0001/fk0002 on A+B (620 bp, EcoRI/BamHI)
Digest pBjk2741-Bca1144 (EcoRI/BamHI, 2170+910, L)
Product is pBjk2741-ssb21 {<FokI+>}
---------------------------------
fk0001 Forward EcoRI and BglII for FokI ccaaaGAATTCgtccAGATCTCAGCTGGTTaaatctgaactggaggag
fk0004 (F)Base Substitution FokI ccataaaaccaatTgcaatggcgccg
fk0003 (R)Base Substitution FokI cggcgccattgcAattggttttatgg
fk0002 Reverse BamHI for FokI gctagGGATCCaaaattgatctccccattgttg
Construction file for sbb08
EIPCR tn0001/tn0002R on pBjk2741-Bca1144 (2230 bp, BglII)
Product is pBjk2741-ssb08 {Tn5 5’TR}
-------
tn0001 EIPCR construction of Tn5 part
CCATAagatctGTCGACTGTCTCTTATACACATCTCAACCggatcctaaCTCGCTCCTCaggcttc
tn0002R Reverse BglII oligo for Tn5 EIPCR
CCAATAGATCTcatgaattcCACTTCAG
17:25, 17 February 2010
Today I set up all my 1st step PCRs for sbb21 and ssb08. I labeled the PCR tubes TN0001/0002R, fk 0001/0003 and fk0004/0002.
The PCR procedure is as follows:
1.Dilute Oligos to 100uM (move decimal point for concentration over by one and add that much ddH2O) 2.Make a seperate dilution of 10uM by adding 1uL of the stock to 9uL of water. Mix well. 3.In a PCR tube add 24uL of ddH2O, 3.3uL of 10x expand buffer and dNTPs, 1uL of each of the respective oligos, 0.5uL Template and LASTLY 0.5 uL of the polymerase.
14:30, 22 February 2010
Today I prepped TN0001/0002R for an analytical gel and FK0001/0003 and FK0004/0002 for the preparative gel.
Lane 10 is TN0001/0002R
Lane 6 is FK 0001/0003 and Lane 7 is FK0002/0004
Analytical gel for TN looked good, so proceeded to zymo clean up.
Ran regular zymo clean up for TN0001/0002R using the following procedure:
1. Add 180uL of Zymo ADB buffer to the 33uL sample of TN 2. Transfered to zymo column 3. Spin in centrifuge at max speed for 30s. Discard the waste. 4. Add 200uL of the wash buffer and spin through. 5. Repeat 6. Spin for 90s to dry 7. Elute with 33uL of elution buffer.
-Stored TN elution in Box A
The preparative gel for Fk 0001/0003 showed the correct size. I cut it from the gel and melted it in 600uL ADB buffer at 55 degrees.
Fk0002/0004 was shown to be a larger size than expected. I repeated the PCR with and with out DMSO. Hopefully, I will get the desired product.
14:12, 24 February 2010
First I prepped the FK0002/0004 with and without DMSO samples for the preparative gel. I then proceeded to prep my TN0001/0002R for the digestion by adding 8.5uL of eluted PCR product to 1uL of NEB Buffer 2 and lastly adding the enzyme 0.5uL BglII. I incubated the samples at 37 degrees for an hour on the thermocycler.
After incubation, I Prepped the BglII digested TN1/2R for a preparative gel. The band looked good, so I proceeded to cut the DNA from the gel and melt it in ADB buffer.
14:30, 26 February 2010
Unfortunately, the FK 0001/0003 and FK 0002/0004 was lost. I then remade the FK0001/0003 and FK0002/0004 oligos, ran preparative gels for each, cut out the segments and melted them in ADB buffer.
14:30, 1 March 2010
Today, I prepped the TN1/2R for and EIPCR ligation and transformation.
For the EIPCR ligation I added the following together in a tube and pounded it on the bench top to mix.
- 7.5uL ddH2O
- 1uL T4 DNA Ligase Buffer (small red or black-striped tubes)
- 1uL PCR product digest
- 0.5uL T4 DNA Ligase
I then placed it in the 37 degree incubator for 30min.
After 30 minutes, I placed the ligation mixture on ice and began transformation.
I first added 50uL of water and 30ul of KCM to a thawed 200uL aliquot of cells. I then added 70uL of this mixture to my ligation and placed it on ice for 10 minutes. I heat shocked the cells for 90 seconds at 42 degrees and placed it back on the ice for 1 minute. Next, I added 100uL of 2YT, being careful to use a flame to keep things sterile, and let the whole mixture shake in the incubator for an hour.
Because I had class at 4PM, Will and Mike plated the bacteria for me.
14:30, 3 March 2010
Today, I miniprepped TN1/2R. After finishing, I continued to prep FK 1/3 2/4 for analytical gel.
The sbb21 (FK 1/3 2/4) product is expected to have a band at ~600bp. The product is in lane 4 and looks approximately at 600bp. Therefore the product is good. I will continue with zymo clean up and proceed with anEcoI/BamHI digestion next week.
14:00, 8 March 2010
Prepared the TN1/2R for mapping. During 30min incubation, I proceeded to zymo the ssb21 PCR product. Accidentally, eluted the DNA into A4 buffer waste. Professor Anderson saved the product by repeating the zymo cleanup and eluting with less water in case some DNA was lost. I then ran a preparative gel on the sample and a band appeared at ~600 as expected. I cut the DNA out of the gel, melted it at 55 degrees, and stored it away to zymo gel purify next week.
After the incubation for TN1/2R, I proceeded to run an analytical gel for all four clones. The result is as follows, where lane 5,6,7,8 correspond to clones 1,2,3,4 respectively.
Clone 3 differs from the others, therefore it may be bad. I chose clone 1 and 2 for sequencing.
14:23, 10 March 2010
Entered clone 1 and 2 for TN1/2R into clotho to get ready for sequencing.
Zymo gel purified FK and proceeded to ligation and transformation.
13:50, 15 March 2010
Transformation failed for sbb21 from last week. I discovered it was because I used the wrong vector plasmid (it wasn't treated with the restriction enzymes).
I redid the ligation transformation for sbb21.
Checked sequencing for sbb08. The part is perfect.
13:30, 17 March 2010
The transformation was successful; however, accidentally no colonies were picked. Will is going to pick colonies today and will continue the mini prep.
12:00, 18 March 2010
Will miniprepped and mapped the clones for sbb23. The results of the gel are as follows:
The size of the digestion should have been around 600bps; however, the gel shows bands at ~3000-4000bps. Clearly the bands are much longer than they should be. Professor Anderson believes something definitely went wrong and suggested we scrap this part and not bother sequencing it. He is going to make a new one from another version of the part that was successful during the break.