SBB10Ntbk-JingLuo: Difference between revisions
No edit summary |
(→3/17) |
||
| (44 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<pre> | |||
Construction of sbb20: FokI Cleavage Domain - <FokI-! | |||
PCR gh1000F/gh1001R on BBa_K243001 (261 bp, gp = A) | |||
PCR gh1001F/gh1003R on BBa_K243001 (264 bp, gp = B) | |||
PCR gh1003F/gh1000R on BBa_K243001 (144 bp, gp = C) | |||
--------------------------------------------------- | |||
PCR gh1000F/gh1000R on A+B+C (622 bp, EcoRI/BamHI) | |||
Digest pBjk2741 (EcoRI/BamHI, 2170+910 L) | |||
Product is pBjk2741-sbb20 | |||
--------------------------------------------------- | |||
gh1000F Forward for part <FokI-! ccagtGAATTCatgAGATCTCAGCTGGTTaaatctgaactggaggag | |||
gh1001F Making internal mutation 1+2 cgttggttccccgatcgattatggcgttatcgtggAcacaaaagc | |||
gh1001R Making internal mutation 1+2 cgccataatcgatcggggaaccaacggtataaatggcaccGTctggtttac | |||
gh1003F Making internal mutation 3 ccatatcaccaatTgcaatggggcagtgctgag | |||
gh1003R Making internal mutation 3 ccattgcAattggtgatatgg | |||
gh1000R Reverse for part <FokI-! gcaaaGGATCCTTAaaaattgatctcgccattgttg | |||
Part: GATCTCAGCTGGTTaaatctgaactggaggagaaaaaatccgagctgcgccacaaactgaaatatgtgcctcacgagtatatcgaactgatcgagatcgcccgtaatagtacccaagaccgtatcctggaaatgaaagtgatggagttcttcatgaaagtctatggctatcgtggcaaacatctgggtggtagccGTAAACCAGACGGTGCCATTTATACcgttggttccccgatcgattatggcgttatcgtggAcacaaaagcgtattctggcggttataatctgccgattggtcaggctgatgagatggaacgttatgtggaagagaatcagacccgtaacaaacatctgaacccgaacgaatggtggaaagtgtatccgtcaagtgtcaccgagttcaaatttctgttcgtgagcggccactttaaaggcaactataaagcccagctgactcgtctgaaccatatcaccaatTgcaatggggcagtgctgagtgttgaggaactgctgatcggtggagaaatgatcaaagcaggcaccctgactctggaagaagttcgccgtaaattCAACAATGGCGAGATCAATTTTTAAG | |||
</pre> | |||
<pre> | |||
Construction of sbb35: P9 rep Protein - {rbs.ColE2 RepA} | |||
Part already exists, only requires a EcoR1/BamH1 transfer into pBca9145-Bca1144. | |||
</pre> | |||
Gel ladder that was used for all the experiments | |||
<br> | |||
[[Image:Generuler1kbplus.jpg]] | |||
<br> | |||
General Experiment Outline: | |||
* Generate the insert | |||
* Restriction enzyme digest the insert and vector | |||
* Purify necessary fragments using preparative gel and zymo gel clean | |||
* Ligate insert and vector together | |||
* Transform with correct cells | |||
* Plate onto appropriate selective antibiotic media | |||
* Pick colonies and culture | |||
* Miniprep colonies | |||
* Analytical digest mapping | |||
* Sequencing of colony that looked correct via digest mapping | |||
Inventory (Inside Box C) | Inventory (Inside Box C) | ||
* y A(star) - part A for SOEing | * y A(star) - part A for SOEing | ||
| Line 9: | Line 50: | ||
* sb20 digest zc - Digested w/ EcoR1/BamH1 and zymo cleaned of the Round 2 SOEing reaction | * sb20 digest zc - Digested w/ EcoR1/BamH1 and zymo cleaned of the Round 2 SOEing reaction | ||
* 20 digest prep gel - sbb20 from preparative gel after digestion | * 20 digest prep gel - sbb20 from preparative gel after digestion | ||
* Xfer digest zc - Eco/Bam transfer digest and zymo cleaned | * Xfer digest zc - Eco/Bam transfer digest and zymo cleaned | ||
* MP20 #1 - Miniprepped sbb20 colony from transformed cells w/ part | * MP20 #1 - Miniprepped sbb20 colony from transformed cells w/ part | ||
| Line 22: | Line 60: | ||
* sb35 4 - Miniprepped sbb35 colony from transformed cells w/ part | * sb35 4 - Miniprepped sbb35 colony from transformed cells w/ part | ||
==2/17== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' Using the designed oligos to clone the fragments of sbb20, which will later be SOE'd together. | <br>'''Purpose:''' Create sbb20 insert -- Using the designed oligos to clone the fragments of sbb20, which will later be SOE'd together. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Using the 2K55 temperature program, cloned the necessary parts for SOEing using the protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_PCR2 Cloning by PCR] | * Using the 2K55 temperature program, cloned the necessary parts for SOEing using the protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_PCR2 Cloning by PCR] | ||
| Line 32: | Line 70: | ||
<br>- y A(star) (sbb20A), y B(star) (sbb20B), y C(star) (sbb20C) | <br>- y A(star) (sbb20A), y B(star) (sbb20B), y C(star) (sbb20C) | ||
==2/22== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' | <br>'''Purpose:''' Create sbb20 insert -- Identify the presence or not of the appropriate sequence, extracting it and purifying it from the gel. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ran a preparative gel on the PCR products from 2/17 (sbb20A,sbb20B,sbb20C) | * Ran a preparative gel on the PCR products from 2/17 (sbb20A,sbb20B,sbb20C) | ||
'''Results''' | '''Results''' | ||
* Results of the preparative gel looked good. Lanes 9, 10, 11 (Part A, B, C respectively): [http://openwetware.org/wiki/Image:XYL022210gel1.jpg Gel pic] | * Results of the preparative gel looked good. Lanes 9, 10, 11 (Part A, B, C respectively): [http://openwetware.org/wiki/Image:XYL022210gel1.jpg Gel pic] | ||
* part a = 261 bp | |||
* part b = 264 bp | |||
* part c = 144 bp | |||
[[Image:XYL022210gel1.jpg]] | |||
<br>Lanes | |||
#ladder | |||
#sbb04A | |||
#sbb04B | |||
#sbb04C | |||
#RH 33A | |||
#RH33B | |||
#blank | |||
#blank | |||
#CD1 <-- Part A | |||
#CD2 <-- Part B | |||
#CD3 <-- Part C | |||
#sbb19A | |||
#sbb19B | |||
#ladder | |||
* Ran a gel purification of the preparative gel fragments using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Zymo3 Zymo gel purification] | |||
'''Products labeled:''' | '''Products labeled:''' | ||
<br>- Preparative gel: JHL Gel frag | <br>- Preparative gel: JHL Gel frag | ||
<br>- Post zymo clean: | <br>- Post zymo clean: PCR Zc ABC | ||
==2/24== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' Using the external primers to combine the three parts together into sbb20. | <br>'''Purpose:''' Create sbb20 insert -- Using the external primers to combine the three parts together into sbb20. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ran a SOEing reaction using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_PCR3 SOEing by PCR] <br> | * Ran a SOEing reaction using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_PCR3 SOEing by PCR] <br> | ||
| Line 52: | Line 109: | ||
* Next step is to run an analytical gel to determine the results of the SOEing reaction. | * Next step is to run an analytical gel to determine the results of the SOEing reaction. | ||
'''Products labeled:''' | '''Products labeled:''' | ||
<br>- | <br>- sbb20 sew | ||
==3/1== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' Identify whether or not the SOEing reaction produced the full length sequence. | <br>'''Purpose:''' Create sbb20 insert -- Identify whether or not the SOEing reaction produced the full length sequence. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ran an analytical gel | * Ran an analytical gel | ||
'''Results''' | '''Results''' | ||
* Results of analytical gel showed SOEing PCR failed to produce any product: [http://openwetware.org/wiki/SBB10_gels#Michel_Nofal_4 Gel pic] | * Results of analytical gel showed SOEing PCR failed to produce any product: [http://openwetware.org/wiki/SBB10_gels#Michel_Nofal_4 Gel pic] | ||
* Full SOEing sequence = 622 bp | |||
[[Image:MN030110.jpg]] | |||
<br> | |||
#Ladder | |||
#RH33-2K45 | |||
#RH33-2K45 w/ DMSO | |||
#sbb02 | |||
#sbb01 | |||
#sbb03 | |||
#JL20 <-- SOEing reaction | |||
#PiggyBacA | |||
#Ladder | |||
* Reattempting the SOEing, by repeating with a modified SOEing PCR to use DMSO or H2O and temperature program 45: [http://openwetware.org/wiki/Template:SBB-Protocols_PCR3 SOEing by PCR] | * Reattempting the SOEing, by repeating with a modified SOEing PCR to use DMSO or H2O and temperature program 45: [http://openwetware.org/wiki/Template:SBB-Protocols_PCR3 SOEing by PCR] | ||
'''Products labeled:''' | '''Products labeled:''' | ||
<br>- sbb20 SOE H2O | <br>- sbb20 SOE H2O | ||
<br>- sbb20 SOE DMSO | <br>- sbb20 SOE DMSO | ||
==3/3== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' Identify the presence or not of the appropriate sequence from the SOEing reactions and extract it. | <br>'''Purpose:''' Create sbb20 insert -- Identify the presence or not of the appropriate sequence from the SOEing reactions and extract it. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ran on preparative gel. | * Ran on preparative gel. | ||
'''Results''' | '''Results''' | ||
* The full length sequence was present. It was extracted and run through gel purification using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Zymo3 Zymo gel purification]. | * The full length sequence was present: [http://openwetware.org/wiki/SBB10_gels#Amy_Kristofferson Gel pic] | ||
* Full SOEing sequence = 622 bp | |||
[[Image:Ak3-3-10.png]] | |||
<br> | |||
#Ladder | |||
#sbb20 Digest <-- sbb20 SOE H2O | |||
#sbb20 Digest <-- sbb20 SOE DMSO | |||
#KRM27 1 | |||
#KRM27 2 | |||
#KRM27 3 | |||
#KRM27 4 | |||
#KRM16 1 | |||
#KRM16 2 | |||
#KRM16 3 | |||
#KRM16 4 | |||
#FK114 | |||
#Ladder | |||
* It was extracted and run through gel purification using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Zymo3 Zymo gel purification]. | |||
* Digested the product with restriction enzymes using the protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz2 EcoR1/BamH1 Digest of PCR products] | * Digested the product with restriction enzymes using the protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz2 EcoR1/BamH1 Digest of PCR products] | ||
'''Products labeled:''' | '''Products labeled:''' | ||
<br>- 20 digest prep gel | <br>- 20 digest prep gel | ||
==3/8== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' Combine the digested insert (sbb20 part) with the vector, transform into cells, and | <br>'''Purpose:''' Create the full correct plasmid (sbb20 part + vector) -- Combine the digested insert (sbb20 part) with the vector, transform into cells, and plate. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ligated the part with the pBjk2741 vector using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz4 Ligation of EcoR1/BamH1 Digests] | * Ligated the part with the pBjk2741 vector using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz4 Ligation of EcoR1/BamH1 Digests] | ||
| Line 87: | Line 176: | ||
Part sbb35: | Part sbb35: | ||
<br>'''Purpose:''' Transfer the part from one vector to another, starting with digestion of the sbb35 part. | <br>'''Purpose:''' Create the full correct plasmid (sbb35 part + vector) -- Transfer the part from one vector to another, starting with digestion of the sbb35 part. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Began a Eco/Bam transfer using protocol: [http://openwetware.org/wiki/Template:SBB-EcoBamXfers EcoR1/BamH1 Part Transfer] | * Began a Eco/Bam transfer using protocol: [http://openwetware.org/wiki/Template:SBB-EcoBamXfers EcoR1/BamH1 Part Transfer] | ||
| Line 96: | Line 185: | ||
'''Products labeled:''' | '''Products labeled:''' | ||
<br>- Xfer digest zc | <br>- Xfer digest zc | ||
==3/9== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' Extract the DNA from the colonies so that it can be analyzed. | <br>'''Results:''' | ||
* Colonies did grow and the GSI's picked out colonies and cultured them using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro2 Picking of Colonies] | |||
==3/10== | |||
Part sbb20: | |||
<br>'''Purpose:''' Create the full correct plasmid (sbb20 part + vector) -- Extract the DNA from the colonies so that it can be analyzed. | |||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Minipepped cells using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro4 Macherey-Nagel Minipepped] | * Minipepped cells using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro4 Macherey-Nagel Minipepped] | ||
'''Results''' | |||
* Need to determine whether or not the cells successfully took the full plasmid, this will be done next time by analytical gel. | |||
'''Products labeled:''' | |||
<br>- MP20 #1, MP20 #2, MP20 #3, and MP20 #4 | |||
Part sbb35: | Part sbb35: | ||
<br>'''Purpose:''' | <br>'''Purpose:''' Create the full correct plasmid (sbb35 part + vector) -- Combine the insert with the vector, transform into cells, and plate. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* The pBca1256 vector was pre-digested by the GSI's using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz2 EcoR1/BamH1 Digest of PCR products] | |||
* Ligated the part with pBca1256 vector instead of pBca9523 using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz4 Ligation of EcoR1/BamH1 Digests] | * Ligated the part with pBca1256 vector instead of pBca9523 using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz4 Ligation of EcoR1/BamH1 Digests] | ||
* Transformed into DH108 spec cells using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro1 Transformation by Heat Shock] | * Transformed into DH108 spec cells using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro1 Transformation by Heat Shock] | ||
* Plated out onto spec antibiotic plate | * Plated out onto spec antibiotic plate | ||
'''Results''' | |||
* The next step is to see if any colonies grow up on the selective resistance plates, if so pick colonies and culture them. | |||
'''Products labeled:''' | |||
<br>- JHL sbb35 | |||
==3/11-15== | |||
Part sbb35: | |||
* GSI's picked a colony and cultured it according to protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro2 Picking of Colonies] | |||
==3/15== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' | <br>'''Purpose:''' Create the full correct plasmid (sbb20 part + vector) -- Determine if the transformed cells DNA contain the full plasmid. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ran an analytical | * Ran an analytical gel with a portion of the miniprepped DNA using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 Analytical digest mapping] | ||
* Results of the analytical | '''Results''' | ||
<br> | * Results of the analytical gel showed that it succeeded. The cells have an approximately full length plasmid: [http://openwetware.org/wiki/SBB10_gels#Jing_Luo Gel pic] | ||
* Insert and vector = 622 bp + 2170 bp | |||
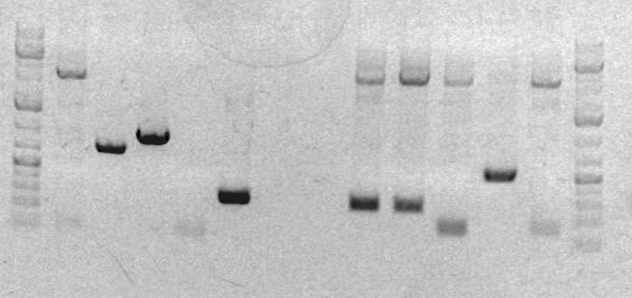
[[Image:Jing gel2.jpg]] | |||
<br> | |||
#Ladder | |||
#sbb36 Clone 1 | |||
#sbb36 Clone 3 | |||
#sbb20 Clone 1 <-- | |||
#sbb20 Clone 2 <-- | |||
#sbb20 Clone 3 <-- | |||
#sbb20 Clone 4 <-- | |||
#Ladder | |||
* The next step is to sequence the plasmid to make sure that it is 100% what we wanted and no mutations. | |||
Part sbb35: | Part sbb35: | ||
<br>'''Purpose:''' | <br>'''Purpose:''' Create the full correct plasmid (sbb35 part + vector) -- Extract and purify the DNA from the cells | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* | * The cells were miniprepped using protocol: [http://openwetware.org/wiki/Template:SBB-Protocols_Micro4 Macherey-Nagel Minipepped] | ||
'''Results''' | |||
* The next step is to run the DNA on an analytical gel, this will allow us to see roughly whether or not the full sequence is present. | |||
'''Products labeled:''' | |||
<br>- sb35 1, sb35 2, sb35 3, sb35 4 | |||
==3/17== | |||
Part sbb20: | Part sbb20: | ||
<br>'''Purpose:''' | <br>'''Purpose:''' Create the full correct plasmid (sbb20 part + vector) -- Check sequence of plasmid to target plasmid. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Sequencing | * Sequencing | ||
'''Results''' | |||
* Will receive the results later. | |||
* Edit later time: Sequence for [Plate A:B6] has numerous mutations and it not usable at all. See [https://spreadsheets.google.com/ccc?key=0AtJsaKPNcPQxdEYyVnZILWRWa0hDQkdldGxrNE1Db3c&hl=en Sequence log] | |||
'''Products labeled:''' | |||
<br>- Took the B6 well for sequencing of the sbb20 | |||
Part sbb35: | Part sbb35: | ||
<br>'''Purpose:''' | <br>'''Purpose:''' Create the full correct plasmid (sbb35 part + vector) -- Determine if the full sequence is present. | ||
<br>'''Protocol:''' | <br>'''Protocol:''' | ||
* Ran an analytical | * Ran an analytical gel | ||
<br> | * The result of the analytical gel showed that it failed. The plasmids turned out to be super huge: [http://openwetware.org/wiki/SBB10_gels#Jeni_Lee2 Gel pic 1+2] [http://openwetware.org/wiki/SBB10_gels#Maziar_Behtash Gel pic 3+4] | ||
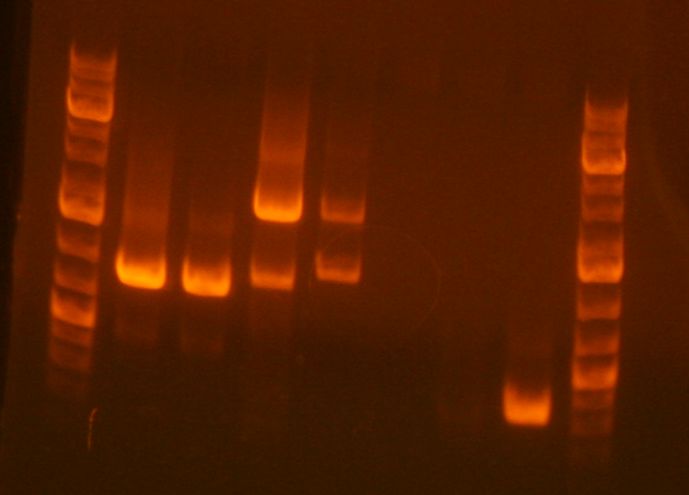
[[Image:Imageprepgeljhl.jpg]] | |||
<br> | |||
#Ladder | |||
#sbb07 digest | |||
#JL clone 1 <-- | |||
#JL clone 2 <-- | |||
[[Image:Mb 03 17 10.jpg]] | |||
<br> | |||
#ladder | |||
#JL3 <-- | |||
#JL4 <-- | |||
#empty | |||
#sbb23-1 | |||
#sbb23-3 | |||
#sbb23-4 | |||
#ladder | |||
* Ran out of time to finish this part and is left as is. | |||
== Team 4: Toxicity and Expression == | |||
[[http://openwetware.org/wiki/SBB10AssayTeam4-Notes Link to Team Notebook]] | |||
Latest revision as of 23:58, 3 May 2010
Construction of sbb20: FokI Cleavage Domain - <FokI-! PCR gh1000F/gh1001R on BBa_K243001 (261 bp, gp = A) PCR gh1001F/gh1003R on BBa_K243001 (264 bp, gp = B) PCR gh1003F/gh1000R on BBa_K243001 (144 bp, gp = C) --------------------------------------------------- PCR gh1000F/gh1000R on A+B+C (622 bp, EcoRI/BamHI) Digest pBjk2741 (EcoRI/BamHI, 2170+910 L) Product is pBjk2741-sbb20 --------------------------------------------------- gh1000F Forward for part <FokI-! ccagtGAATTCatgAGATCTCAGCTGGTTaaatctgaactggaggag gh1001F Making internal mutation 1+2 cgttggttccccgatcgattatggcgttatcgtggAcacaaaagc gh1001R Making internal mutation 1+2 cgccataatcgatcggggaaccaacggtataaatggcaccGTctggtttac gh1003F Making internal mutation 3 ccatatcaccaatTgcaatggggcagtgctgag gh1003R Making internal mutation 3 ccattgcAattggtgatatgg gh1000R Reverse for part <FokI-! gcaaaGGATCCTTAaaaattgatctcgccattgttg Part: GATCTCAGCTGGTTaaatctgaactggaggagaaaaaatccgagctgcgccacaaactgaaatatgtgcctcacgagtatatcgaactgatcgagatcgcccgtaatagtacccaagaccgtatcctggaaatgaaagtgatggagttcttcatgaaagtctatggctatcgtggcaaacatctgggtggtagccGTAAACCAGACGGTGCCATTTATACcgttggttccccgatcgattatggcgttatcgtggAcacaaaagcgtattctggcggttataatctgccgattggtcaggctgatgagatggaacgttatgtggaagagaatcagacccgtaacaaacatctgaacccgaacgaatggtggaaagtgtatccgtcaagtgtcaccgagttcaaatttctgttcgtgagcggccactttaaaggcaactataaagcccagctgactcgtctgaaccatatcaccaatTgcaatggggcagtgctgagtgttgaggaactgctgatcggtggagaaatgatcaaagcaggcaccctgactctggaagaagttcgccgtaaattCAACAATGGCGAGATCAATTTTTAAG
Construction of sbb35: P9 rep Protein - {rbs.ColE2 RepA}
Part already exists, only requires a EcoR1/BamH1 transfer into pBca9145-Bca1144.
Gel ladder that was used for all the experiments
General Experiment Outline:
- Generate the insert
- Restriction enzyme digest the insert and vector
- Purify necessary fragments using preparative gel and zymo gel clean
- Ligate insert and vector together
- Transform with correct cells
- Plate onto appropriate selective antibiotic media
- Pick colonies and culture
- Miniprep colonies
- Analytical digest mapping
- Sequencing of colony that looked correct via digest mapping
Inventory (Inside Box C)
- y A(star) - part A for SOEing
- y B(star) - part B for SOEing
- y C(star) - part C for SOEing
- JHL Gel frag - preparative gel fragments of PCR A, B, C dissolved in ADB buffer
- PCR Zc ABC - Zymo gel clean preparative gel fragments of PCR A, B, C
- sbb20 Sew - SOEing with external primers, first attempt
- sbb20 SOE H2O - 2nd round of SOEing of PCR products A, B, C
- sb20 digest zc - Digested w/ EcoR1/BamH1 and zymo cleaned of the Round 2 SOEing reaction
- 20 digest prep gel - sbb20 from preparative gel after digestion
- Xfer digest zc - Eco/Bam transfer digest and zymo cleaned
- MP20 #1 - Miniprepped sbb20 colony from transformed cells w/ part
- MP20 #2 - Miniprepped sbb20 colony from transformed cells w/ part
- MP20 #3 - Miniprepped sbb20 colony from transformed cells w/ part
- MP20 #4 - Miniprepped sbb20 colony from transformed cells w/ part
- sb35 1 - Miniprepped sbb35 colony from transformed cells w/ part
- sb35 2 - Miniprepped sbb35 colony from transformed cells w/ part
- sb35 3 - Miniprepped sbb35 colony from transformed cells w/ part
- sb35 4 - Miniprepped sbb35 colony from transformed cells w/ part
2/17
Part sbb20:
Purpose: Create sbb20 insert -- Using the designed oligos to clone the fragments of sbb20, which will later be SOE'd together.
Protocol:
- Using the 2K55 temperature program, cloned the necessary parts for SOEing using the protocol: Cloning by PCR
Results
- The next step is to run a preparative gel to determine the results of the PCR.
Products labeled:
- y A(star) (sbb20A), y B(star) (sbb20B), y C(star) (sbb20C)
2/22
Part sbb20:
Purpose: Create sbb20 insert -- Identify the presence or not of the appropriate sequence, extracting it and purifying it from the gel.
Protocol:
- Ran a preparative gel on the PCR products from 2/17 (sbb20A,sbb20B,sbb20C)
Results
- Results of the preparative gel looked good. Lanes 9, 10, 11 (Part A, B, C respectively): Gel pic
- part a = 261 bp
- part b = 264 bp
- part c = 144 bp
- ladder
- sbb04A
- sbb04B
- sbb04C
- RH 33A
- RH33B
- blank
- blank
- CD1 <-- Part A
- CD2 <-- Part B
- CD3 <-- Part C
- sbb19A
- sbb19B
- ladder
- Ran a gel purification of the preparative gel fragments using protocol: Zymo gel purification
Products labeled:
- Preparative gel: JHL Gel frag
- Post zymo clean: PCR Zc ABC
2/24
Part sbb20:
Purpose: Create sbb20 insert -- Using the external primers to combine the three parts together into sbb20.
Protocol:
- Ran a SOEing reaction using protocol: SOEing by PCR
Results
- Next step is to run an analytical gel to determine the results of the SOEing reaction.
Products labeled:
- sbb20 sew
3/1
Part sbb20:
Purpose: Create sbb20 insert -- Identify whether or not the SOEing reaction produced the full length sequence.
Protocol:
- Ran an analytical gel
Results
- Results of analytical gel showed SOEing PCR failed to produce any product: Gel pic
- Full SOEing sequence = 622 bp
- Ladder
- RH33-2K45
- RH33-2K45 w/ DMSO
- sbb02
- sbb01
- sbb03
- JL20 <-- SOEing reaction
- PiggyBacA
- Ladder
- Reattempting the SOEing, by repeating with a modified SOEing PCR to use DMSO or H2O and temperature program 45: SOEing by PCR
Products labeled:
- sbb20 SOE H2O
- sbb20 SOE DMSO
3/3
Part sbb20:
Purpose: Create sbb20 insert -- Identify the presence or not of the appropriate sequence from the SOEing reactions and extract it.
Protocol:
- Ran on preparative gel.
Results
- The full length sequence was present: Gel pic
- Full SOEing sequence = 622 bp
- Ladder
- sbb20 Digest <-- sbb20 SOE H2O
- sbb20 Digest <-- sbb20 SOE DMSO
- KRM27 1
- KRM27 2
- KRM27 3
- KRM27 4
- KRM16 1
- KRM16 2
- KRM16 3
- KRM16 4
- FK114
- Ladder
- It was extracted and run through gel purification using protocol: Zymo gel purification.
- Digested the product with restriction enzymes using the protocol: EcoR1/BamH1 Digest of PCR products
Products labeled:
- 20 digest prep gel
3/8
Part sbb20:
Purpose: Create the full correct plasmid (sbb20 part + vector) -- Combine the digested insert (sbb20 part) with the vector, transform into cells, and plate.
Protocol:
- Ligated the part with the pBjk2741 vector using protocol: Ligation of EcoR1/BamH1 Digests
- Transformed into JTK086 spec cells using protocol: Transformation by Heat Shock
- GSI's plated out onto spec antibiotic plate.
Results
- The next step is to see if any colonies grow up on the selective resistance plates, if so pick colonies and culture them.
Products labeled:
- JHL sbb20 plate
Part sbb35:
Purpose: Create the full correct plasmid (sbb35 part + vector) -- Transfer the part from one vector to another, starting with digestion of the sbb35 part.
Protocol:
- Began a Eco/Bam transfer using protocol: EcoR1/BamH1 Part Transfer
- stopped after the restriction enzyme digestion that was done using protocol: EcoR1/BamH1 Digest of PCR products
- performed a regular zymo clean up on the part using protocol: Regular zymo cleanup
Results
- The next step is to digest the vector and ligate the two together, whether or not this works will be determined once the cells are miniprepped and a analytical digest is run.
Products labeled:
- Xfer digest zc
3/9
Part sbb20:
Results:
- Colonies did grow and the GSI's picked out colonies and cultured them using protocol: Picking of Colonies
3/10
Part sbb20:
Purpose: Create the full correct plasmid (sbb20 part + vector) -- Extract the DNA from the colonies so that it can be analyzed.
Protocol:
- Minipepped cells using protocol: Macherey-Nagel Minipepped
Results
- Need to determine whether or not the cells successfully took the full plasmid, this will be done next time by analytical gel.
Products labeled:
- MP20 #1, MP20 #2, MP20 #3, and MP20 #4
Part sbb35:
Purpose: Create the full correct plasmid (sbb35 part + vector) -- Combine the insert with the vector, transform into cells, and plate.
Protocol:
- The pBca1256 vector was pre-digested by the GSI's using protocol: EcoR1/BamH1 Digest of PCR products
- Ligated the part with pBca1256 vector instead of pBca9523 using protocol: Ligation of EcoR1/BamH1 Digests
- Transformed into DH108 spec cells using protocol: Transformation by Heat Shock
- Plated out onto spec antibiotic plate
Results
- The next step is to see if any colonies grow up on the selective resistance plates, if so pick colonies and culture them.
Products labeled:
- JHL sbb35
3/11-15
Part sbb35:
- GSI's picked a colony and cultured it according to protocol: Picking of Colonies
3/15
Part sbb20:
Purpose: Create the full correct plasmid (sbb20 part + vector) -- Determine if the transformed cells DNA contain the full plasmid.
Protocol:
- Ran an analytical gel with a portion of the miniprepped DNA using protocol: Analytical digest mapping
Results
- Results of the analytical gel showed that it succeeded. The cells have an approximately full length plasmid: Gel pic
- Insert and vector = 622 bp + 2170 bp
- Ladder
- sbb36 Clone 1
- sbb36 Clone 3
- sbb20 Clone 1 <--
- sbb20 Clone 2 <--
- sbb20 Clone 3 <--
- sbb20 Clone 4 <--
- Ladder
- The next step is to sequence the plasmid to make sure that it is 100% what we wanted and no mutations.
Part sbb35:
Purpose: Create the full correct plasmid (sbb35 part + vector) -- Extract and purify the DNA from the cells
Protocol:
- The cells were miniprepped using protocol: Macherey-Nagel Minipepped
Results
- The next step is to run the DNA on an analytical gel, this will allow us to see roughly whether or not the full sequence is present.
Products labeled:
- sb35 1, sb35 2, sb35 3, sb35 4
3/17
Part sbb20:
Purpose: Create the full correct plasmid (sbb20 part + vector) -- Check sequence of plasmid to target plasmid.
Protocol:
- Sequencing
Results
- Will receive the results later.
- Edit later time: Sequence for [Plate A:B6] has numerous mutations and it not usable at all. See Sequence log
Products labeled:
- Took the B6 well for sequencing of the sbb20
Part sbb35:
Purpose: Create the full correct plasmid (sbb35 part + vector) -- Determine if the full sequence is present.
Protocol:
- Ran an analytical gel
- The result of the analytical gel showed that it failed. The plasmids turned out to be super huge: Gel pic 1+2 Gel pic 3+4
- Ladder
- sbb07 digest
- JL clone 1 <--
- JL clone 2 <--
- ladder
- JL3 <--
- JL4 <--
- empty
- sbb23-1
- sbb23-3
- sbb23-4
- ladder
- Ran out of time to finish this part and is left as is.