SBB10Ntbk-DorothyTulanont: Difference between revisions
| Line 162: | Line 162: | ||
Need to make new construction files for Sbb25 and sbb26. | Need to make new construction files for Sbb25 and sbb26. | ||
==[[User:Dorothy D. Tulanont | Dorothy Tulanont]] | ==[[User:Dorothy D. Tulanont | Dorothy Tulanont]] 12 March 2010== | ||
New construction files | New construction files | ||
| Line 188: | Line 188: | ||
I will also be doing | I will also be doing | ||
[http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 Analytical digests (Mapping)] of all my 8 tubes. Since both sbb25 and sbb26 parts are greater than 250bp, I can use EcoRI and BamHI restriction enzyme for my analytical digests. | [http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 Analytical digests (Mapping)] of all my 8 tubes. Since both sbb25 and sbb26 parts are greater than 250bp, I can use EcoRI and BamHI restriction enzyme for my analytical digests. | ||
=Expected Product from EcoRI/BamHI restriction= | |||
pBca1256-sbb25: 645+2472 | |||
pBca1256-sbb26: 671+2472 | |||
<pre> | <pre> | ||
Revision as of 09:16, 12 March 2010
Main page
Protocol
General Idea:
PCR, analytical gel, zymo clean up, digestion, gel purificatoin, ligation, transformation, pick colonies, miniprep, mapping
Dorothy Tulanont 1 February 2010
Today, I received my project ball.
Project Description
Dorothy Tulanont 17 February 2010
Construction File of sbb25 PCR sbb25-F and sbb25-R on pEC22-CA42 (661 bp, EcoRI/BamHI) Sub into pBca9523-Bca1455 (EcoRI/BamHI, 2472+915, L) Product is pBca9523-sbb25 ---------------------------- sbb25-F Cloning of sbb25 ccataGAATTCatgAGATCTcagctgaagtgaccggattagc sbb25-R Cloning of sbb25 ctgatGGATCCgcacgccctttgccggagatgcgaag
Construction File of sbb26 PCR sbb26-F and sbb26-R on pEC22-P9 (687 bp, EcoRI/BamHI) Sub into pBca9523-Bca1455 (EcoRI/BamHI, 2472+915, L) Product is pBca9523-sbb26 ---------------------------- sbb26-F Cloning of sbb26 ccataGAATTCatgAGATCTtgaagtgaccggattagcaacgc sbb26-R Cloning of sbb26 ctgatGGATCCcaatgtttttcggcagaatgcgaacgccg
Construction File of Pre-Pro sequence sbb43 (not PCR)
sbb43 (109bp when cut with EcoRI and BamHI)
Digest pBjk2741 (EcoRI/BamHI, 2170+910, L)
Product is pBjk2741-Bjh1730 {rbs2_lamB>}
I prepared sbb25 and sbb26 for PCR reaction using cloning by PCR protocol. I also threw the diluted primer DNA (10uM) away.
Since both of my PCR products are less than 2kb (SBB25 = 661bp and SBB26 = 687bp), the PCR reaction will run in 2K55 mode.
Dorothy Tulanont 22 February 2010
Today, I ran analytical gel on my sbb25 and sbb26 products by mixing 3uL of the product and 7uL of the loading dye.
This is the result of my analytical gel 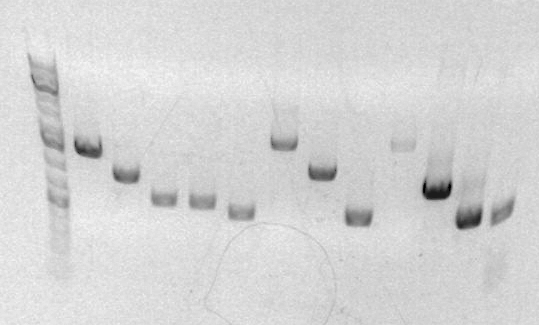
After my parts were verified, I did Zymo clean up on my PCR product.
Lane 1 = Ladder Lane 2, 3 = Jenna (SBB28, SBB29) Lane 4, 5 = Dorothy (SBB25 SBB26) Lane 6 = Carolyn Lane 7 = Jeni (SBB34) Lane 8 = Jose (JG1) Lane 9 = ZZH Lane 10 = Paulina (Th0001//0002R)
My tubes are labeled "SBB25 DDT (Zymo Done)" and "SBB26 DDT (Zymo Done)." These tubes are then stored in 140L box A.
Dorothy Tulanont 24 February 2010
Today, I did EcoRI/BamHI Digest of both of my PCR products (sbb25 and sbb26). My tubes are labeled "sbb25" and "sbb26" respectively. Then I did preparative gel on both of my digests (SBB25 and SBB26).
Dorothy Tulanont 1 March 2010
Today, I started with Eco/Bam Transfer for sbb43: Pre-Pro sequence. While I was waiting for the digestion, I did Zymo Gel Purification for SBB25 and SBB26. The tubes are labeled "SBB25 Zymo GP" and "SBB26 Zymo GP" respectively. They are stored in box A along with "SBB25 DDT (Zymo Done)" and "SBB26 DDT (Zymo Done)."
SBB 43
Source: pBjh1601AC-Bjh1730
Vector: pBjk2741 (resistant to spectinomycin)
Competent Cells: JTK086 (double blue stripes)
Dorothy Tulanont 3 March 2010
Today, I did minipreping on my sbb43 cell culture. Four different colonies were minipreped. The tubes are labeled "SBB43-A" "SBB43-B" "SBB43-C" "SBB43-D" respectively.
On Monday, I will run Analytical digests (Mapping) gel for "SBB43-A" "SBB43-B" "SBB43-C" "SBB43-D" respectively which are stored in miniprep box. Also, I will do ligation and transformation for sbb25, sbb26.
Dorothy Tulanont 8 March 2010
Sbb43 (A, B, C, D)
Progress: Digest, ligation, transformation, pick colonies, miniprep.
Need to do: Analytical digests (Mapping)
Sbb 25, 26
Progress: PCR, analytical gel, zymo clean up, digestion, gel purification
Need to do: ligation, transformation, pick colonies, miniprep, Analytical digests (mapping)
Today
"SBB25 Zymo GP" and "SBB26 Zymo GP"
ligation and transformation
Ligation of EcoRI/BamHI digests *Set up the following reaction: 6.5uL ddH2O 1uL T4 DNA Ligase Buffer (small red or black-striped tubes) 1uL Vector digest (pBca9523, use pBca1256 instead) 1uL Insert digest 0.5uL T4 DNA Ligase *Pound upside down on the bench to mix *Give it a quick spin to send it back to the bottom of the tube *Incubate on the benchtop for 30min (2:01PM) *Put on ice and proceed to the transformation
Transformation by heat-shock '''Competent cells are stored as 200uL aliquots in the -80 freezer as a communal stock.''' 1. Thaw a 200 uL aliquot of cells on ice 2. Add 50 uL of water to the cells 3. Add 30 uL of KCM to the cells 4. Put your ligation mixture on ice, let cool a minute or two 5. Add 70 uL of the cell cocktail to the ligation, stir to mix 6. Let sit on ice for 10 min (2:35PM) 7. Heat shock for 90 seconds at 42 (longer incubation may work better) 8. Put back on ice for 1 min 9. Add 100uL of 2YT, let shake in the 37 degree incubator for 1 hour 10. Plate 70+ uL on selective antibiotics, let incubate at 37 degrees overnight
"SBB43-A" "SBB43-B" "SBB43-C" "SBB43-D"
Analytical digests (Mapping)
Since the part is only 109bp (&2170bp) long when cut with EcoRI and BamHI, I will cut my miniprepped plasmid with EcoRI and XhoI (1595 + 684)
Set up the following 10uL reaction in a PCR tube: 4uL ddH2O 4uL Miniprepped plasmid 1uL 10x NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI (for parts >250bp) or XhoI (for parts <250bp) *Incubate at 37 on the thermocycler for 30 minutes (2:30PM) *Run an analytical gel *Take a picture of the gel *Calculate the expected fragment sizes *Are the calculated sizes consistent with the bands on the gel? YES!
This is a picture of my analytical gel for mapping. Lane 2-5. The parts should be 1595 and 684


Note: pBca9523 was not used for Sbb24 and Sbb25. I used pBca1256 instead. I also accidentally added 70uL of cells into my zymo gel purification Sbb25.
Dorothy Tulanont 10 March 2010
Today, I will miniprep cells that was plated and cultured since 3/8/10 (Sbb25 and sbb26). There will be microcentrifuge tubes labeled "Sbb25-A, Sbb25-B, Sbb25-C, Sbb25-D" and "Sbb26-A, Sbb26-B, Sbb26-C, Sbb26-D" (8 tubes total).
I am going to send Sbb43A and Sbb43B to sequence. The information are put into clotho
Note: The plate for Sbb25 and Sbb26 were contaminated. Hopefully the right E.Coli was picked and cultured.
All minipreped DNA are stored in the miniprepped box. (2-9, 17, 18, 26, 27)
Need to make new construction files for Sbb25 and sbb26.
Dorothy Tulanont 12 March 2010
New construction files
Construction File of sbb25 PCR sbb25-F and sbb25-R on pEC22-CA42 (661 bp, EcoRI/BamHI) Sub into pBca1256 (EcoRI/BamHI, 63 + 2481 , L) Product is pBca1256-sbb25 ---------------------------- sbb25-F Cloning of sbb25 ccataGAATTCatgAGATCTcagctgaagtgaccggattagc sbb25-R Cloning of sbb25 ctgatGGATCCgcacgccctttgccggagatgcgaag
Construction File of sbb26 PCR sbb26-F and sbb26-R on pEC22-P9 (687 bp, EcoRI/BamHI) Sub into pBca1256 (EcoRI/BamHI, 36+2481 , L) Product is pBca1256-sbb26 ---------------------------- sbb26-F Cloning of sbb26 ccataGAATTCatgAGATCTtgaagtgaccggattagcaacgc sbb26-R Cloning of sbb26 ctgatGGATCCcaatgtttttcggcagaatgcgaacgccg
I will also be doing
Analytical digests (Mapping) of all my 8 tubes. Since both sbb25 and sbb26 parts are greater than 250bp, I can use EcoRI and BamHI restriction enzyme for my analytical digests.
Expected Product from EcoRI/BamHI restriction
pBca1256-sbb25: 645+2472 pBca1256-sbb26: 671+2472
Set up the following 10uL reaction in a PCR tube: 4uL ddH2O 4uL Miniprepped plasmid 1uL 10x NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI *Incubate at 37 on the thermocycler for 30 minutes (Time: ?? PM) *Run an analytical gel *Take a picture of the gel *Calculate the expected fragment sizes *Are the calculated sizes consistent with the bands on the gel?