SBB10Ntbk-DorothyTulanont: Difference between revisions
No edit summary |
|||
| Line 78: | Line 78: | ||
Need to do: Analytical digests (Mapping) <br> | Need to do: Analytical digests (Mapping) <br> | ||
<br> | <br> | ||
Set up the following 10uL reaction in a PCR tube: | |||
<pre> | |||
4uL ddH2O | |||
4uL Miniprepped plasmid | |||
1uL 10x NEB Buffer 2 | |||
0.5uL EcoRI | |||
0.5uL BamHI (for parts >250bp) or XhoI (for parts <250bp) | |||
</pre> | |||
*Incubate at 37 on the thermocycler for 30 minutes | |||
*Run an analytical gel | |||
*Take a picture of the gel | |||
*Calculate the expected fragment sizes | |||
*Are the calculated sizes consistent with the bands on the gel? | |||
<br> | |||
Sbb 25, 26<br> | Sbb 25, 26<br> | ||
Progress: PCR, analytical gel, zymo clean up, digestion, gel purification <br> | Progress: PCR, analytical gel, zymo clean up, digestion, gel purification <br> | ||
Revision as of 13:55, 8 March 2010
Main page
Protocol
General Idea:
PCR, analytical gel, zymo clean up, digestion, gel purificatoin, ligation, transformation, pick colonies, miniprep, mapping
Dorothy Tulanont 1 February 2010
Today, I received my project ball.
Project Description
Dorothy Tulanont 17 February 2010
Construction File of sbb25 PCR sbb25-F and sbb25-R on pEC22-CA42 (661 bp, EcoRI/BamHI) Sub into pBca9523-Bca1455 (EcoRI/BamHI, 2472+915, L) Product is pBca9523-sbb25 ---------------------------- sbb25-F Cloning of sbb25 ccataGAATTCatgAGATCTcagctgaagtgaccggattagc sbb25-R Cloning of sbb25 ctgatGGATCCgcacgccctttgccggagatgcgaag
Construction File of sbb26 PCR sbb26-F and sbb26-R on pEC22-P9 (687 bp, EcoRI/BamHI) Sub into pBca9523-Bca1455 (EcoRI/BamHI, 2472+915, L) Product is pBca9523-sbb26 ---------------------------- sbb26-F Cloning of sbb26 ccataGAATTCatgAGATCTtgaagtgaccggattagcaacgc sbb26-R Cloning of sbb26 ctgatGGATCCcaatgtttttcggcagaatgcgaacgccg
I prepared sbb25 and sbb26 for PCR reaction using cloning by PCR protocol. I also threw the diluted primer DNA (10uM) away. Since both of my PCR products are less than 2kb (SBB25 = 661bp and SBB26 = 687bp), the PCR reaction will run in 2K55 mode.
Dorothy Tulanont 22 February 2010
Today, I ran analytical gel on my sbb25 and sbb26 products by mixing 3uL of the product and 7uL of the loading dye.
This is the result of my analytical gel 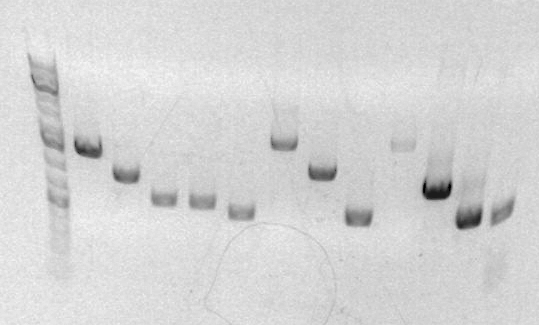
After my parts were verified, I did Zymo clean up on my PCR product.
Lane 1 = Ladder Lane 2, 3 = Jenna (SBB28, SBB29) Lane 4, 5 = Dorothy (SBB25 SBB26) Lane 6 = Carolyn Lane 7 = Jeni (SBB34) Lane 8 = Jose (JG1) Lane 9 = ZZH Lane 10 = Paulina (Th0001//0002R)
My tubes are labeled "SBB25 DDT (Zymo Done)" and "SBB26 DDT (Zymo Done)." These tubes are then stored in 140L box A.
Dorothy Tulanont 24 February 2010
Today, I did EcoRI/BamHI Digest of both of my PCR products (sbb25 and sbb26). My tubes are labeled "sbb25" and "sbb26" respectively. Then I did preparative gel on both of my digests (SBB25 and SBB26).
Dorothy Tulanont 1 March 2010
Today, I started with Eco/Bam Transfer for sbb43: Pre-Pro sequence. While I was waiting for the digestion, I did Zymo Gel Purification for SBB25 and SBB26. The tubes are labeled "SBB25 Zymo GP" and "SBB26 Zymo GP" respectively. They are stored in box A along with "SBB25 DDT (Zymo Done)" and "SBB26 DDT (Zymo Done)."
SBB 43
Source: pBjh1601AC-Bjh1730
Vector: pBjk2741 (resistant to spectinomycin)
Competent Cells: JTK086 (double blue stripes)
Dorothy Tulanont 3 March 2010
Today, I did minipreping on my sbb43 cell culture. Four different colonies were minipreped. The tubes are labeled "SBB43-A" "SBB43-B" "SBB43-C" "SBB43-D" respectively.
On Monday, I will run Analytical digests (Mapping) gel for "SBB43-A" "SBB43-B" "SBB43-C" "SBB43-D" respectively which are stored in miniprep box. Also, I will do ligation and transformation for sbb25, sbb26.
Dorothy Tulanont 8 March 2010
Sbb43 (A, B, C, D)
Progress: Digest, ligation, transformation, pick colonies, miniprep.
Need to do: Analytical digests (Mapping)
Set up the following 10uL reaction in a PCR tube:
4uL ddH2O 4uL Miniprepped plasmid 1uL 10x NEB Buffer 2 0.5uL EcoRI 0.5uL BamHI (for parts >250bp) or XhoI (for parts <250bp)
- Incubate at 37 on the thermocycler for 30 minutes
- Run an analytical gel
- Take a picture of the gel
- Calculate the expected fragment sizes
- Are the calculated sizes consistent with the bands on the gel?
Sbb 25, 26
Progress: PCR, analytical gel, zymo clean up, digestion, gel purification
Need to do: ligation, transformation, pick colonies, miniprep, Analytical digests (mapping)
Today
"SBB43-A" "SBB43-B" "SBB43-C" "SBB43-D"
Analytical digests (Mapping)
"SBB25 Zymo GP" and "SBB26 Zymo GP"
ligation and transformation