SBB10Ntbk-AmyKristofferson: Difference between revisions
| (29 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[http://openwetware.org/wiki/SynBioBootcamp:Notebook/SynBioBootcamp10 BioE140L 2010 Homepage]<br> | [http://openwetware.org/wiki/SynBioBootcamp:Notebook/SynBioBootcamp10 BioE140L 2010 Homepage]<br> | ||
[http://openwetware.org/wiki/Template:SBB10Project-19244 My Project Page]<br> | [http://openwetware.org/wiki/Template:SBB10Project-19244 My Project Page]<br> | ||
==3/20/10: Success!== | |||
*My sequencing results are perfect. The part sbb22 was a success! | |||
==3/17/10: Submitted sbb22 for Sequencing== | |||
*I logged my samples into clotho and loaded sbb22-1 and sbb22-2 for sequencing. | |||
==3/15/10== | |||
===sbb22 Wetlab notes: Mapping attempt #2=== | |||
*I mapped my plasmids again and got the following analytical gel: | |||
[[Image:WJH_gels_031510.jpg]]<br> | |||
My lanes are as follows:<br> | |||
6. sbb22-1 <br> | |||
7. sbb22-2 <br> | |||
8. sbb22-3 <br> | |||
9. sbb22-4 <br> | |||
I'm going to have 1 and 2 sequenced, since they both appear to have a band around 600bp. There are two bands between 2000 and 3000bp, which is probably due to an incomplete digestion. | |||
===sbb36 Wetlab notes: Miniprep, Mapping, and Failure=== | |||
*The GSI's [http://openwetware.org/wiki/Template:SBB-Protocols_Micro2 picked colonies] for me, so today I performed a [http://openwetware.org/wiki/Template:SBB-Protocols_Micro4 mini prep]. I labeled the tubes sbb36 plasmid 1-4, where the numbers correspond to the well I removed the cells from. Only tubes 1 and 3 produced a pellet. | |||
*I also [http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 mapped] the plasmids. Sbb36-1 is in lane 2 and sbb36-3 is in lane 3. Since nothing substantial turned up, I'm not going to sequence any plasmids for this part and assume that the project failed. Prof Anderson will redo it over Spring Break. | |||
[[Image:Jing_gel2.jpg]] | |||
==3/10/10== | ==3/10/10== | ||
===sbb22 Wetlab notes=== | ===sbb22 Wetlab notes: Miniprep and unsuccessful Mapping=== | ||
*The GSI's [http://openwetware.org/wiki/Template:SBB-Protocols_Micro2 picked colonies] for me, so today I performed a [http://openwetware.org/wiki/Template:SBB-Protocols_Micro4 mini prep]. I labeled the tubes sbb22 plasmid 1-4, where the numbers correspond to the well I removed the cells from. There could be complications though, because the media was contaminated. Along with several fairly large, translucent white colonies (there was only one red colony), there were a ton of tiny white colonies- the contaminent. | *The GSI's [http://openwetware.org/wiki/Template:SBB-Protocols_Micro2 picked colonies] for me, so today I performed a [http://openwetware.org/wiki/Template:SBB-Protocols_Micro4 mini prep]. I labeled the tubes sbb22 plasmid 1-4, where the numbers correspond to the well I removed the cells from. There could be complications though, because the media was contaminated. Along with several fairly large, translucent white colonies (there was only one red colony), there were a ton of tiny white colonies- the contaminent. | ||
*I also [http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 mapped] the plasmids. | *I also [http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 mapped] the plasmids. | ||
The four lanes on the right are of my part. My part should be 603bp long, but there's no band of that length in any of my lanes. If the plasmid had just closed on itself, I would only expect a band at 2170bp. It appears that this was the case; that I was just unlucky enough to choose four colonies that picked up plasmids that didn't contain my part. However, I'm going to map my mini-prepped plasmids again to make sure that I just didn't make a mistake when I digested them when prepping for the analytical gel. | |||
[[Image:Mb031010.jpg]] | [[Image:Mb031010.jpg]] | ||
===sbb36 Wetlab notes: Ligation and Transformation=== | |||
===sbb36 Wetlab notes=== | |||
*[http://openwetware.org/wiki/Template:SBB-Protocols_Enz4 Ligation] of digest product and pre-digested pBca1256-Bca1144(instead of pBca9523- Bca1144, which I originally said I would use in my construction file) | *[http://openwetware.org/wiki/Template:SBB-Protocols_Enz4 Ligation] of digest product and pre-digested pBca1256-Bca1144(instead of pBca9523- Bca1144, which I originally said I would use in my construction file) | ||
*[http://openwetware.org/wiki/Template:SBB-Protocols_Micro1 Transformation] | *[http://openwetware.org/wiki/Template:SBB-Protocols_Micro1 Transformation] into strain DH10B | ||
==3/8/10 | ==3/8/10== | ||
===sbb22 Wetlab notes=== | ===sbb22 Wetlab notes: Ligation and Transformation=== | ||
*[http://openwetware.org/wiki/Template:SBB-Protocols_Zymo3 Gel purification] of Digested sbb22 PCR product | *[http://openwetware.org/wiki/Template:SBB-Protocols_Zymo3 Gel purification] of Digested sbb22 PCR product | ||
| Line 20: | Line 43: | ||
*[http://openwetware.org/wiki/Template:SBB-Protocols_Micro1 Transformation] (I used the JTK086 cell strain) | *[http://openwetware.org/wiki/Template:SBB-Protocols_Micro1 Transformation] (I used the JTK086 cell strain) | ||
===sbb36 Wetlab notes=== | ===sbb36 Wetlab notes: Digest=== | ||
'''Eco/Bam Transfer''' <br> | '''Eco/Bam Transfer''' <br> | ||
(I amended [http://openwetware.org/wiki/Template:SBB-EcoBamXfers this protocol] so I could digest today and then ligate and transform on Wednesday) | (I amended [http://openwetware.org/wiki/Template:SBB-EcoBamXfers this protocol] so I could digest today and then ligate and transform on Wednesday) | ||
| Line 41: | Line 64: | ||
*Incubate 30 min. at room temp, transform, rescue, plate. | *Incubate 30 min. at room temp, transform, rescue, plate. | ||
==3/3/10: Successful SOEing PCR and Eco/Bam Digest | ==3/3/10== | ||
===sbb22 Wetlab notes: Successful SOEing PCR and Eco/Bam Digest=== | |||
*Ran an analytical gel of two PCR products | *Ran an analytical gel of two PCR products | ||
[[Image:3-3-10-gel.png]]<br> | [[Image:3-3-10-gel.png]]<br> | ||
| Line 56: | Line 79: | ||
My digest is in lane 2. | My digest is in lane 2. | ||
==3/1/10: Trying out alternative SOEing PCR techniques== | ==3/1/10== | ||
===sbb22 Wetlab notes: Trying out alternative SOEing PCR techniques=== | |||
I'm going to redo the SOEing PCR (using oligos gh1000F/ak11R on A+B+C template) using the 2K45 PCR program, since my first attempt with 55 failed. | I'm going to redo the SOEing PCR (using oligos gh1000F/ak11R on A+B+C template) using the 2K45 PCR program, since my first attempt with 55 failed. | ||
I'll set up one tube following the original protocol, and then another one with 3.3 uL of DMSO substituted for the usual 3.3uL of ddH2O. | I'll set up one tube following the original protocol, and then another one with 3.3 uL of DMSO substituted for the usual 3.3uL of ddH2O. | ||
| Line 64: | Line 88: | ||
I labeled the PCR tubes "sbb22 H2O" and "sbb22 DMSO." | I labeled the PCR tubes "sbb22 H2O" and "sbb22 DMSO." | ||
==2/24/10 | ==2/24/10== | ||
===Wetlab Notes=== | ===sbb22 Wetlab Notes: SOEing PCR and Analytical Gel=== | ||
*PCR gh1000F/ak11R on A+B+C using [http://openwetware.org/wiki/Template:SBB-Protocols_PCR2 this protocol] and 55 PCR. | *PCR gh1000F/ak11R on A+B+C using [http://openwetware.org/wiki/Template:SBB-Protocols_PCR2 this protocol] and 55 PCR. | ||
*Ran analytical gel to verify that desired PCR product was created. (Protocol: add 3uL PCR product and 7uL of loading buffer) | *Ran analytical gel to verify that desired PCR product was created. (Protocol: add 3uL PCR product and 7uL of loading buffer) | ||
| Line 73: | Line 97: | ||
*While waiting for the gel, I performed a Zymo clean-up to remove dNTPs, Expand Polymerase, and oligos. What's left: the <500bp PCR product, A, B, C. DNA labeled "sbb22, final PCR product, AK" and placed in Box A. (3-3-10: I'm not using this PCR product, so I threw it out) | *While waiting for the gel, I performed a Zymo clean-up to remove dNTPs, Expand Polymerase, and oligos. What's left: the <500bp PCR product, A, B, C. DNA labeled "sbb22, final PCR product, AK" and placed in Box A. (3-3-10: I'm not using this PCR product, so I threw it out) | ||
===Protocol Update=== | ===sbb22 Protocol Update=== | ||
Next few steps (after performing a successful SOEing PCR) | Next few steps (after performing a successful SOEing PCR) | ||
*Eco/Bam Digest of PCR product (vector has already been digested) | *Eco/Bam Digest of PCR product (vector has already been digested) | ||
| Line 79: | Line 103: | ||
*Cut out digested product and perform a Gel purification | *Cut out digested product and perform a Gel purification | ||
==2/22/10: Preparative Gel for PCR product A,B,C== | ==2/22/10== | ||
===sbb22 Wetlab Notes: Preparative Gel for PCR product A,B,C=== | |||
Mixed 6uL of each PCR product with 4uL of loading buffer and ran the gel. | Mixed 6uL of each PCR product with 4uL of loading buffer and ran the gel. | ||
| Line 89: | Line 113: | ||
I cut out the bottom bands (the upper band is most likely template DNA), performed a small fragment Zymo Cleanup, and stored the eluted DNA in Box A. | I cut out the bottom bands (the upper band is most likely template DNA), performed a small fragment Zymo Cleanup, and stored the eluted DNA in Box A. | ||
==2/17/10 | ==2/17/10== | ||
===New sbb22 Construction File=== | ===New sbb22 Construction File=== | ||
<pre> | <pre> | ||
| Line 118: | Line 142: | ||
*Set up your second round of PCR as a normal 33uL reaction using the eluted mixture of fragments as template | *Set up your second round of PCR as a normal 33uL reaction using the eluted mixture of fragments as template | ||
=== | ===sbb22 Wetlab notes: PCR of A,B,C off of template=== | ||
Prepared oligos and set up three PCR tubes using [http://openwetware.org/wiki/Template:SBB-Protocols_PCR2 this protocol]. | Prepared oligos and set up three PCR tubes using [http://openwetware.org/wiki/Template:SBB-Protocols_PCR2 this protocol]. | ||
The PCR tubes were labeled A, B, and C and placed in Box A. | The PCR tubes were labeled A, B, and C and placed in Box A. | ||
Latest revision as of 10:03, 3 May 2010
BioE140L 2010 Homepage
My Project Page
3/20/10: Success!
- My sequencing results are perfect. The part sbb22 was a success!
3/17/10: Submitted sbb22 for Sequencing
- I logged my samples into clotho and loaded sbb22-1 and sbb22-2 for sequencing.
3/15/10
sbb22 Wetlab notes: Mapping attempt #2
- I mapped my plasmids again and got the following analytical gel:

My lanes are as follows:
6. sbb22-1
7. sbb22-2
8. sbb22-3
9. sbb22-4
I'm going to have 1 and 2 sequenced, since they both appear to have a band around 600bp. There are two bands between 2000 and 3000bp, which is probably due to an incomplete digestion.
sbb36 Wetlab notes: Miniprep, Mapping, and Failure
- The GSI's picked colonies for me, so today I performed a mini prep. I labeled the tubes sbb36 plasmid 1-4, where the numbers correspond to the well I removed the cells from. Only tubes 1 and 3 produced a pellet.
- I also mapped the plasmids. Sbb36-1 is in lane 2 and sbb36-3 is in lane 3. Since nothing substantial turned up, I'm not going to sequence any plasmids for this part and assume that the project failed. Prof Anderson will redo it over Spring Break.
3/10/10
sbb22 Wetlab notes: Miniprep and unsuccessful Mapping
- The GSI's picked colonies for me, so today I performed a mini prep. I labeled the tubes sbb22 plasmid 1-4, where the numbers correspond to the well I removed the cells from. There could be complications though, because the media was contaminated. Along with several fairly large, translucent white colonies (there was only one red colony), there were a ton of tiny white colonies- the contaminent.
- I also mapped the plasmids.
The four lanes on the right are of my part. My part should be 603bp long, but there's no band of that length in any of my lanes. If the plasmid had just closed on itself, I would only expect a band at 2170bp. It appears that this was the case; that I was just unlucky enough to choose four colonies that picked up plasmids that didn't contain my part. However, I'm going to map my mini-prepped plasmids again to make sure that I just didn't make a mistake when I digested them when prepping for the analytical gel.
sbb36 Wetlab notes: Ligation and Transformation
- Ligation of digest product and pre-digested pBca1256-Bca1144(instead of pBca9523- Bca1144, which I originally said I would use in my construction file)
- Transformation into strain DH10B
3/8/10
sbb22 Wetlab notes: Ligation and Transformation
- Gel purification of Digested sbb22 PCR product
- Ligation of sbb22 and pre-digested pBjk2741-Bca1144
- Transformation (I used the JTK086 cell strain)
sbb36 Wetlab notes: Digest
Eco/Bam Transfer
(I amended this protocol so I could digest today and then ligate and transform on Wednesday)
- Set up the digest:
For BBb transfers: 6 uL ddH2O (changed from 7.5uL) 1 uL NEB2+ATP Buffer 0.5uL EcoRI 0.5uL BamHI 2 uL miniprepped plasmid: pBca9145-jtk2642 (changed from 0.5uL)
- Incubate at 37 degrees on the thermocycler for 1hr
- Perform Zymo clean-up. Now I should have purified insert and old plasmid DNA.
Note: a preparative gel isn't necessary to separate the insert from the original vector, because I will be able to select for the bacteria with the new vector by plating on spec, since the new vector has a specR gene, while the old vector does not. I labeled the tube "sbb36 Digest AK" and put it in Box A.
And then next time...
- Add 0.5uL of predigest vector and 0.5uL of T4 DNA ligase
- Incubate 30 min. at room temp, transform, rescue, plate.
3/3/10
sbb22 Wetlab notes: Successful SOEing PCR and Eco/Bam Digest
- Ran an analytical gel of two PCR products

Lane 2: sbb22 H2O
Lane 3: sbb22 DMSO
Both PCR products contained a band around 600bp, but the band in lane 2 was darker, so I'm going to use "sbb22 H2O" to continue on with my project. I labeled it "sbb22 H2O AK" and put it in box A. I threw out the "sbb22 DMSO" product.
- Performed a Zymo Cleanup of products while waiting for results.
- Performed a Eco/Bam digest on my "sbb22 H2O" PCR product using this protocol.
- Ran a preparative gel of digest, cut out higher band and melted gel in AB buffer.
3/1/10
sbb22 Wetlab notes: Trying out alternative SOEing PCR techniques
I'm going to redo the SOEing PCR (using oligos gh1000F/ak11R on A+B+C template) using the 2K45 PCR program, since my first attempt with 55 failed. I'll set up one tube following the original protocol, and then another one with 3.3 uL of DMSO substituted for the usual 3.3uL of ddH2O.
I labeled the PCR tubes "sbb22 H2O" and "sbb22 DMSO."
2/24/10
sbb22 Wetlab Notes: SOEing PCR and Analytical Gel
- PCR gh1000F/ak11R on A+B+C using this protocol and 55 PCR.
- Ran analytical gel to verify that desired PCR product was created. (Protocol: add 3uL PCR product and 7uL of loading buffer)

My PCR product was labeled sbb22 and was in the fourth lane from the left. The PCR product should have been 619bp, but all the bands that showed up were under 500 bps.
- While waiting for the gel, I performed a Zymo clean-up to remove dNTPs, Expand Polymerase, and oligos. What's left: the <500bp PCR product, A, B, C. DNA labeled "sbb22, final PCR product, AK" and placed in Box A. (3-3-10: I'm not using this PCR product, so I threw it out)
sbb22 Protocol Update
Next few steps (after performing a successful SOEing PCR)
- Eco/Bam Digest of PCR product (vector has already been digested)
- Run a preparative gel to separate digested product from A, B, and C and to ensure product was correctly digested
- Cut out digested product and perform a Gel purification
2/22/10
sbb22 Wetlab Notes: Preparative Gel for PCR product A,B,C
Mixed 6uL of each PCR product with 4uL of loading buffer and ran the gel.

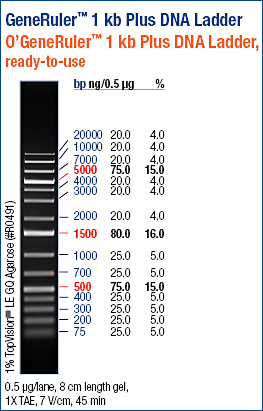
My PCR products A, B, and C were in lanes 2, 3, and 4.
I cut out the bottom bands (the upper band is most likely template DNA), performed a small fragment Zymo Cleanup, and stored the eluted DNA in Box A.
2/17/10
New sbb22 Construction File
Construction of FokI- basic part
PCR gh1000F/gh1001R on BBa_K243001 (261 bp, gp = A)
PCR gh1001F/gh1003R on BBa_K243001 (264 bp, gp = B)
PCR gh1003F/ak11R Registry part BBa_K243001 (141 bp, gp = C)
---------------------------------------------------
PCR gh1000F/ak11R on A+B+C (619bp, EcoRI/BamHI)
Digest pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L)
Product is pBjk2741-sbb22 {<FokI->}
---------------------------------------------------
gh1000F Forward for part <FokI-! ccagtGAATTCatgAGATCTCAGCTGGTTaaatctgaactggaggag
gh1001F Making internal mutation 1+2 cgttggttccccgatcgattatggcgttatcgtggAcacaaaagc
gh1001R Making internal mutation 1+2 cgccataatcgatcggggaaccaacggtataaatggcaccGTctggtttac
gh1003F Making internal mutation 3 ccatatcaccaatTgcaatggggcagtgctgag
gh1003R Making internal mutation 3 ccattgcAattggtgatatgg
ak11R Reverse BamHI for FokI- CTGATggatccaaaattgatctcgccattg
SOEing Protocol for sbb22
- Set up PCR reactions according to your construction file as normal 33uL reactions as described in Cloning by PCR
- For each PCR, load 6uL of PCR product premixed with 4uL of loading buffer in a single well of a 1% agarose gel
- Cut out the bands, put them into a single 1.5mL microcentrifuge tube
- Add 650uL of ADB Buffer
- Proceed with the Zymo Gel Purification procedure
- Elute the DNA in 50uL of water
- Set up your second round of PCR as a normal 33uL reaction using the eluted mixture of fragments as template
sbb22 Wetlab notes: PCR of A,B,C off of template
Prepared oligos and set up three PCR tubes using this protocol. The PCR tubes were labeled A, B, and C and placed in Box A.
2/8/10: Construction Files for my parts
sbb22
Construction of FokI- basic part
PCR ak10F/ak20R Registry part BBa_K243001 (230 bp, gp = A)
PCR ak20F/ak30R Registry part BBa_K243001 (71 bp, gp = B)
PCR ak30F/ak40R Registry part BBa_K243001 (245 bp, gp = C)
PCR ak40F/ak11R Registry part BBa_K243001 (135 bp, gp = D)
---------------------------------------------------
PCR ak10F/ak11R on A+B+C+D (619bp, EcoRI/BamHI)
Digest pBjk2741-Bca1144 (EcoRI/BamHI, 910+2170, L)
Product is pBjk2741-Bak1234 {<FokI->}
---------------------------------------------------
ak10F Forward EcoRI for FokI- CCATAgaattcCAGagatctCAGCTGGTTaaatctgaactggaggagaa
ak20F Base substitution 1 gtaaaccagACggtgccatt
ak20R Base substitution 1 aatggcaccGTctggtttac
ak30F Base substitution 2 cgttatcgtggAcacaaaagcg
ak30R Base substitution 2 cgcttttgtgTccacgataacgc
ak40F Base substitution 3 caccaatTgcaatggggcag
ak40R Base substitution 3 ctgccccattgcAattggtg
ak11R Reverse BamHI for FokI- CTGATggatccaaaattgatctcgccattg
sbb36
Construction of ColE2 Basic Part
Digest pBca9145-jtk2642 (EcoRI/BamHI, 2057+485, S)
Sub into pBca9523- Bca1144 (EcoRI/BamHI, 2472+918, L)
Product is pBca9523-jtk2642 {ColE2 ori med}


